RNA World Hypothesis and Prebiotic Chemistry: From Life's Origins to Modern Therapeutics
This article explores the RNA World hypothesis, the leading framework for understanding life's origins, and its profound implications for modern biomedical research.
RNA World Hypothesis and Prebiotic Chemistry: From Life's Origins to Modern Therapeutics
Abstract
This article explores the RNA World hypothesis, the leading framework for understanding life's origins, and its profound implications for modern biomedical research. We examine foundational evidence that RNA first catalyzed life's emergence, recent breakthroughs in connecting RNA to amino acids under prebiotic conditions, and methodological advances in RNA engineering. The content addresses key challenges in replicating primordial RNA systems and validates these models with ancient biosignatures and comparative analysis. For researchers and drug development professionals, we synthesize how ancient RNA mechanisms are inspiring revolutionary therapeutics, including RNA-targeting small molecules, mRNA vaccines, and synthetic biological systems.
The RNA World: Foundations of Life and Evolutionary Evidence
Core Principles of the RNA World Hypothesis
The RNA World Hypothesis represents a foundational concept in origins-of-life research, proposing that early life forms relied on RNA for both genetic information storage and catalytic functions before the evolutionary emergence of DNA and proteins. This whitepaper examines the core principles, supporting evidence, methodological approaches, and persistent challenges of this hypothesis within the context of prebiotic chemistry. We provide a technical overview of experimental and computational tools for studying RNA structure and function, highlighting implications for therapeutic development. Despite significant validation from ribozyme discoveries and laboratory evolution experiments, the hypothesis faces challenges regarding RNA's prebiotic synthesis and stability, driving continued interdisciplinary investigation into life's origins.
The RNA World Hypothesis addresses a fundamental paradox in molecular biology: modern cells require proteins to synthesize DNA, yet DNA is essential to code for those same proteins. This interdependence creates a "chicken-and-egg" dilemma regarding which came first in the evolution of life [1] [2]. The hypothesis resolves this by proposing that RNA once served both roles—acting as both the genetic blueprint and the catalytic engine for early life forms [3] [4].
The conceptual foundations were laid in the 1960s by several scientists, including Francis Crick, Carl Woese, and Leslie Orgel [3] [4]. The term "RNA World" itself was later coined by Walter Gilbert in 1986, solidifying the concept within the scientific lexicon [3]. This framework has since become the leading paradigm for understanding the transition from prebiotic chemistry to biological systems, suggesting that around 4 billion years ago, RNA was the primary living substance prior to the evolutionary emergence of DNA-based genomes and protein-based enzymes [3] [2].
Table: Historical Development of the RNA World Hypothesis
| Year | Key Scientist(s) | Contribution |
|---|---|---|
| 1960s | Francis Crick, Carl Woese, Leslie Orgel | Independently proposed core concepts of RNA's dual role |
| 1986 | Walter Gilbert | Coined the term "RNA World" |
| 1989 | Sidney Altman, Thomas Cech | Discovery of catalytic RNA (ribozymes); Nobel Prize in Chemistry |
| 1990s-Present | Various Research Groups | Laboratory demonstrations of RNA self-replication and catalytic diversification |
Core Principles and Theoretical Framework
The RNA World Hypothesis rests on several interconnected principles that collectively describe a plausible pathway for the emergence of life from prebiotic chemistry.
The Dual Functionality of RNA
Unlike DNA, which primarily serves as a passive information repository, RNA can perform two critical functions:
- Genetic Information Storage: RNA can encode hereditary information via its nucleotide sequence, similar to DNA [3] [1].
- Catalytic Activity: RNA can fold into complex three-dimensional structures that form active sites, enabling it to catalyze biochemical reactions much like protein-based enzymes [1] [4]. These catalytic RNA molecules are known as ribozymes.
The Precellular Life Model
Experts generally agree that non-living chemicals could not have directly formed bacterial cells in a single step [3] [4]. The hypothesis posits that self-replicating RNA molecules constituted a critical intermediate, pre-cellular life form. These molecular entities could have carried genetic information across generations independently, undergoing Darwinian evolution before the advent of the modern cell [3].
Resolution of the Biochemical Paradox
The central dogma of molecular biology describes a unidirectional flow of information from DNA → RNA → Protein. However, this system is interdependently complex. The RNA World elegantly resolves this by proposing a simpler, ancestral state where a single polymer type (RNA) handled both informational and functional roles [1] [2]. This period of evolution is thought to have preceded the division of labor that now characterizes biology, where DNA specializes in genetic storage and proteins in catalysis [3].
Evolutionary Transition to DNA and Proteins
The hypothesis does not suggest that the RNA World persists unchanged today. Rather, it proposes that evolution eventually favored DNA for genetic stability and proteins for catalytic efficiency [3]. DNA's double-stranded structure provides greater chemical stability and replication fidelity compared to the more labile RNA [3]. Proteins, with their diverse amino acid side chains, offer a broader range of catalytic activities. The conversion of RNA to DNA via reverse transcription is seen as a key evolutionary step that cemented this transition [3] [4].
Key Evidence and Validation
Substantial experimental and observational evidence has accumulated to support the plausibility of the RNA World Hypothesis.
The Discovery of Ribozymes
The hypothesis gained significant credibility with the landmark discovery that RNA can act as an enzyme. Sidney Altman and Thomas Cech were awarded the 1989 Nobel Prize in Chemistry for their work identifying ribozymes—RNA molecules that catalyze specific biochemical reactions [3] [4]. This shattered the long-held paradigm that only proteins could serve catalytic roles in biology.
The Ribosome as a Ribozyme
Perhaps the most compelling evidence comes from the structure of the ribosome, the cellular machine that synthesizes proteins. High-resolution structural studies reveal that the catalytic peptidyl transferase activity—the formation of peptide bonds between amino acids—is performed by ribosomal RNA (rRNA), not by the ribosomal proteins that provide structural support [3] [1]. This indicates that RNA catalyzes the synthesis of proteins, powerfully supporting the idea that RNA-based catalysis preceded protein-based enzymes.
Laboratory Evolution of Functional RNAs
In vitro evolution experiments have demonstrated that random RNA sequences can evolve to perform diverse functions. Researchers have generated RNA ligases (which join RNA strands) and even RNAs capable of catalyzing limited self-replication from random sequence pools [3] [1]. These findings demonstrate that RNA possesses an inherent functional capacity that could have been exploited by early evolution.
Table: Experimentally Evolved Ribozymes and Their Functions
| Ribozyme Function | Experimental Finding | Significance for RNA World |
|---|---|---|
| Self-Replication | RNA-catalyzed RNA polymerization observed [1] | Demonstrates potential for genetic continuity |
| Amino Acid Ligation | RNA catalysts can join amino acids [1] | Suggests a pathway for early peptide synthesis |
| Peptide Bond Formation | Ribozymes capable of forming peptide bonds [1] | Supports origin of protein synthesis within an RNA framework |
| RNA Ligase Activity | Active ligases derived from random RNA sequences [3] [4] | Shows functional complexity can arise from random sequences |
Methodological Approaches for RNA Structure and Function Analysis
Understanding RNA's catalytic and informational roles requires detailed knowledge of its structure. The following experimental workflows and reagents are fundamental to this research.
RNA Structure Probing with Nucleases
RNA structure can be analyzed using enzymes that cleave RNA at specific sites based on secondary structure. This protocol reveals regions that are single-stranded versus double-stranded [5].
Diagram: RNA Structure Analysis with Nucleases. This workflow uses structure-specific ribonucleases to probe RNA conformation, followed by gel separation to identify cleavage sites.
Table: Research Reagent Solutions for RNA Structure Analysis
| Reagent | Function/Description | Application in Protocol |
|---|---|---|
| End-labeled RNA | RNA labeled with 32P at either 5' (using KinaseMax) or 3' end (using T4 RNA Ligase) | Provides detectable signal for visualization |
| Yeast RNA | Carrier RNA (10 mg/ml) | Stabilizes low-concentration target RNA during processing |
| 10X RNA Structure Buffer | (e.g., 100 mM Tris pH 7, 1 M KCl, 100 mM MgCl2) | Provides optimal ionic conditions for RNA folding |
| RNase T1 | Cleaves 3' of single-stranded G residues | Sequence/structure-specific probing |
| RNase A | Cleaves 3' of single-stranded C and U residues | Sequence/structure-specific probing |
| RNase V1 | Cleaves base-paired nucleotides | Double-stranded region identification |
| Inactivation/Precipitation Buffer | Contains salts and ethanol | Stops reaction and precipitates RNA for cleanup |
| Acrylamide Gel Loading Buffer | (95% Formamide, 18 mM EDTA, 0.025% SDS, dyes) | Denatures RNA for accurate size separation on gel |
High-Throughput RNA Structure Probing
Modern methods couple structure-sensitive chemical probing with high-throughput sequencing to analyze thousands of RNAs simultaneously, creating "structuromes" [6] [7]. These techniques include SHAPE-Seq, DMS-Seq, and SHAPE-MaP, which can be performed both in vitro and in vivo [7].
Diagram: High-Throughput RNA Structure Probing. This conceptual framework underpins transcriptome-wide RNA structure analysis, encoding structural information into cDNA libraries for sequencing.
Table: High-Throughput RNA Structure Probing Techniques
| Method | Probe | Detection Principle | Key Application |
|---|---|---|---|
| SHAPE-Seq | 1M7, BzCN, DMS | Reverse Transcription Stop (RT-Stop) | In vitro/in vivo RNA folding, RNA-ligand interactions |
| DMS-Seq | DMS | RT-Stop | Transcriptome-wide in vivo probing (e.g., yeast, human) |
| SHAPE-MaP | 1M7, 1M6, NMIA, DMS | Reverse Transcription Mutation (RT-Mutate) | Viral genome structures, in vivo lncRNA structures |
| icSHAPE | NAI-N3 | RT-Stop | Transcriptome-wide probing in mouse; effect of RNA modifications |
| CIRS-seq | DMS, CMCT | RT-Stop | Transcriptome-wide in vitro probing in mouse cell lines |
| DMS-MaP-Seq | DMS | RT-Mutate | Global transcriptome analysis in S. cerevisiae, Drosophila |
Challenges and Critical Assessment
Despite its widespread acceptance, the RNA World Hypothesis faces significant challenges that drive ongoing research in prebiotic chemistry and alternative models.
Prebiotic Synthesis and Stability Concerns
A major criticism centers on whether RNA could have formed abiotically on early Earth. Key challenges include:
- Chemical Complexity: RNA is a relatively complex molecule, and the prebiotic pathways for forming its components (ribose sugar, nucleobases, phosphate) and linking them together remain incompletely solved [8].
- Instability: RNA is chemically fragile, particularly the bond between the ribose and nucleobase, which is prone to hydrolysis in water and susceptible to degradation under UV radiation [3] [8]. This raises questions about its persistence in a prebiotic environment.
NASA noted in a 1996 report that "significant difficulties" surrounding the RNA World concept include RNA's chemical fragility and its limited range of catalytic activities compared to proteins [3] [4]. Biochemist Harold S. Bernhardt has pointedly referred to it as "the worst theory of the early evolution of life (except for all the others)" [8], highlighting that while problematic, it remains the most viable framework available.
Limited Catalytic Repertoire
While ribozymes exist, their catalytic efficiency and diversity generally pale in comparison to proteins. Naturally occurring ribozymes primarily catalyze phosphorylation and transesterification reactions involving other RNAs [8]. The hypothesis requires that RNA catalyzed a much broader set of metabolic reactions in the primordial world, a premise that still lacks robust experimental support.
Evolving Terminology in Prebiotic Chemistry
The field is increasingly mindful of terminology. Some researchers suggest that the term "prebiotic chemistry" can be misleading, as it may imply a teleological progression toward life and an over-reliance on traditional chemical synthesis puzzles [9]. Alternative terms like "protobiotic processes" have been proposed to more accurately describe processes assumed to contribute directly to life's emergence, without implying a foreordained outcome [9]. This reflects a more nuanced understanding of the transition from non-living to living matter.
Implications for Biomedical Research and Therapeutics
Understanding RNA's fundamental biology and structural principles, rooted in the RNA World, directly informs modern therapeutic development.
- RNA-Targeted Therapeutics: The development of RNA-based drugs (e.g., mRNA vaccines, RNA interference therapies, riboswitches) relies on precise knowledge of RNA structure-function relationships [6]. High-throughput structure determination methods enable the identification of potential drug targets within RNA genomes of pathogens [6] [7].
- Antibiotic Development: Many antibiotics target the bacterial ribosome. Understanding the ribosome as a ribozyme has been crucial for designing drugs that inhibit its function without affecting host cells [3].
- Synthetic Biology: Researchers are designing novel ribozymes and deoxyribozymes for industrial and diagnostic applications, effectively harnessing the catalytic potential of nucleic acids first proposed by the RNA World Hypothesis [6].
The RNA World Hypothesis remains the most compelling framework for understanding the origin of life, supported by robust evidence from ribozyme biology, structural studies of the ribosome, and laboratory evolution experiments. While legitimate challenges persist regarding the prebiotic synthesis of RNA and its chemical stability, ongoing methodological advances in RNA structure determination and a refining understanding of prebiotic environments continue to address these knowledge gaps. The hypothesis not only provides a historical narrative for life's beginnings but also continues to fuel innovation in biomedical research, particularly in the rapidly expanding field of RNA therapeutics. Future research integrating chemistry, biology, and planetary science will further test the boundaries of this foundational theory.
The ribosome, the universal ribonucleoprotein complex responsible for protein synthesis, provides one of the most compelling molecular fossils for studying the origin and evolution of life. Contemporary structural, phylogenetic, and experimental analyses of ribosomal components offer a window into the prebiotic world and strongly support the RNA world hypothesis, which posits that early life relied on RNA for both genetic information storage and catalytic functions. This technical review synthesizes evidence from structural biology, molecular evolution, and prebiotic chemistry to elucidate how the ribosome's architecture records a chronological evolutionary timeline. We further present quantitative analyses of proto-ribosome emergence probabilities, detailed experimental methodologies for key supporting studies, and essential research tools for investigators in this field.
The RNA world hypothesis represents a dominant paradigm for understanding the origin of life, proposing that RNA-based life forms preceded the DNA/protein world [10] [11]. This hypothesis resolves the fundamental "chicken-and-egg" dilemma of molecular evolution: which came first, proteins that catalyze reactions or DNA that stores genetic information? RNA uniquely addresses this paradox by serving both as a catalyst and an information repository [12]. Within this framework, the ribosome stands as the most significant molecular fossil, providing architectural evidence of a transitional world where RNA catalyzed critical biochemical reactions independently of proteins.
The conclusive evidence emerged from high-resolution ribosome structures, which revealed that the peptidyl transferase center (PTC) – the active site for peptide bond formation – consists exclusively of ribosomal RNA (rRNA) with no proteins in the immediate vicinity [10] [13]. This finding demonstrated that the ribosome is fundamentally a ribozyme, an RNA enzyme that catalyzes the chemical reaction linking amino acids into proteins. The implications are profound: the modern ribosome retains structural features of an ancient RNA machine that likely functioned in the prebiotic era before the emergence of coded protein synthesis.
Structural Evidence from the Contemporary Ribosome
The Ribosomal RNA Core as a Catalytic Center
Seminal biochemical and structural studies have established that rRNA alone catalyzes peptide bond formation. Early reductive experiments by Noller and colleagues demonstrated that the large ribosomal subunit from Thermus aquaticus retained peptidyl transferase activity even after approximately 95% of its protein components were removed [10]. This finding strongly indicated that the 23S rRNA was the catalytic engine. Subsequent high-resolution X-ray crystallography studies solved the complete atomic structure of the 50S ribosomal subunit, providing definitive structural evidence: no protein chains were observed within 18 Å of the PTC, confirming that peptide bond formation is catalyzed solely by rRNA [10].
The structural organization of the modern ribosome further reinforces this conclusion. Contrary to earlier models that viewed rRNA as merely a scaffold for ribosomal proteins, structural analyses reveal that proteins are peripheral components sprinkled across the surface of a massive RNA core that constitutes the ribosome's primary architectural and functional element [13]. This inverted relationship – with RNA forming the catalytic core and proteins playing primarily structural and supportive roles – provides compelling evidence that the ribosome evolved from an earlier RNA-only complex.
Symmetrical Regions and the Proto-Ribosome
Deep within the large ribosomal subunit lies a region of approximately 180 nucleotides exhibiting an approximate 2-fold rotational symmetry [14]. This symmetrical region (SymR), located at the heart of the PTC, is universally conserved across all domains of life (Bacteria, Archaea, and Eukarya) and contains the binding sites for the 3' ends of aminoacyl-tRNA (A-site) and peptidyl-tRNA (P-site) [14]. The symmetry suggests that the modern PTC evolved from the dimerization of two identical or similar RNA molecules that catalyzed primitive peptide bond formation before the emergence of the genetic code.
Research has focused on three concentric structural models of potential proto-ribosomes of dimeric nature:
Table 1: Proto-Ribosome Structural Models
| Model | Size (nucleotides in bacteria) | Structural Description | Key Features |
|---|---|---|---|
| Extended Symmetrical Region (ext-SymR) | 225 | SymR plus non-symmetrical parts of helices H75 and H91 | Largest model; unique pattern of A-minor interactions interpreted as a mode for adding new elements |
| Symmetrical Region (SymR) | 178 | Entire symmetrical region surrounding PTC | Contains the modern peptidyl transferase center; highly conserved structure and sequence |
| Dimeric Proto-Ribosome (DPR) | 121 | Core of SymR; dimer of L-shaped RNA elements | Simplest model; monomers comparable in size/shape to tRNA; most plausible for spontaneous emergence |
These three contenders share a common dimerization mode via GNRA interaction motifs (where N = any nucleotide, R = purine), a known contributor to RNA dimer stability [14]. The DPR model, comprising a dimer of tRNA-like molecules embedded in the core of the symmetrical region, represents the most feasible starting point for continuous evolutionary path from prebiotic chemistry to the modern translation system due to its structural simplicity and higher probability of spontaneous emergence [14].
Evolutionary Chronology Through Structural Analysis
The "Peeling the Onion" Methodology
A groundbreaking approach to establishing ribosomal evolutionary chronologies involves sectioning the large ribosomal subunit (LSU) into concentric shells using the peptidyl transfer site as the origin (PT-origin) [15] [16] [17]. This "peeling the onion" methodology, applied to high-resolution structures from disparate evolutionary lineages (Haloarcula marismortui [archaeal] and Thermus thermophilus [bacterial]), captures significant temporal information by analyzing structural and sequence conservation relative to distance from the catalytic center.
The analysis reveals that sequence and conformational similarity of the 23S rRNAs are greatest near the PT-origin and diverge smoothly with increasing distance from it [15]. This conservation gradient suggests that the ribosome evolved outward from its functional core, with the most ancient components located in the immediate vicinity of the peptidyl transferase site and more recent additions positioned peripherally.
Diagram: Ribosomal Evolution Concentric Shell Model - The "peeling the onion" approach reveals that regions closest to the catalytic center are most ancient and conserved.
Patterns of Molecular Evolution
The concentric shell analysis reveals several fundamental patterns in ribosomal evolution:
RNA Structural Evolution: The tendency of rRNA to assume regular A-form helices with Watson-Crick base pairs is lowest near the PT-origin and increases with distance from it, suggesting that early RNA elements were potentially less structured and may have involved partially single-stranded oligomers assembled with magnesium ion mediation [15] [17].
Protein Recruitment: Ribosomal proteins near the PT-origin are notably shorter in length and display nearly absent secondary structure (α-helices and β-sheets), suggesting they may be molecular fossils of the peptide ancestors of ribosomal proteins [15]. As distance increases from the PT-origin, proteins become larger and incorporate more regular secondary structural elements.
Cofactor Replacement: The early peptidyl transferase center likely relied on Mg²⁺-mediated assembly of RNA components. Moving from center to periphery, proteins appear to progressively replace magnesium ions in structural and potentially catalytic roles [15] [17].
These observable patterns demonstrate that the conformation and interactions of both RNA and protein components change systematically along an evolutionary timeline embedded within the ribosome's architecture.
Probabilistic Assessment of Prebiotic Feasibility
A critical challenge for origin-of-life scenarios is demonstrating the realistic probability that functional proto-molecules could self-assemble from random molecular polymers in prebiotic conditions. For the RNA world hypothesis, this requires assessing the statistical likelihood that a functional proto-ribosome could emerge spontaneously.
Table 2: Probability Analysis of Proto-Ribosome Emergence
| Model | Monomer Length | Probability of Random Sequence | Feasibility Assessment |
|---|---|---|---|
| Extended SymR | 225 nucleotides | Highly implausible (requires ~10¹³⁵ kg RNA) | Not feasible for spontaneous emergence |
| Symmetrical Region | 178 nucleotides | Implausible (requires ~10¹⁰⁶ kg RNA) | Highly unlikely |
| Dimeric Proto-Ribosome | 61-63 nucleotides | Possible with "limited specificity" | The only model with realistic statistical likelihood |
The inverse relationship between ribozyme sequence length and spontaneous emergence probability makes the dimeric nature of the DPR critical to its feasibility. A dimeric structure increases the probability of random emergence by many orders of magnitude compared to a monomer of equivalent total length [14]. Introducing the concept of "limited specificity" – where only a subset of nucleotides must be constrained to preserve structure and function – further enhances the statistical likelihood of DPR emergence from random RNA chains.
For perspective, even a simple 40-nucleotide ribozyme has 4⁴⁰ (approximately 10²⁴) possible sequences. To represent all compositions at least once would require approximately 27 kg of random RNA chains, making spontaneous emergence highly implausible for longer sequences [14]. The DPR, with its shorter monomers and limited specificity requirements, represents the only model with a realistic probability of materializing in prebiotic conditions.
Experimental Protocols and Methodologies
Protocol: Structural Analysis of Ribosomal Evolution
Objective: To establish evolutionary chronologies through comparative analysis of ribosomal large subunit structures.
Materials:
- High-resolution ribosome structures from phylogenetically disparate organisms (e.g., Haloarcula marismortui [PDB: 1FFK] and Thermus thermophilus [PDB: 1GIY])
- Structural superposition software (e.g., PyMOL, Chimera)
- Sequence alignment tools (e.g., ClustalOmega, MUSCLE)
Methodology:
- Structural Superposition: Align large ribosomal subunits using conserved regions around the peptidyl transferase center as reference points.
- Concentric Shell Definition: Define a series of concentric shells radiating outward from the peptidyl transfer origin (PT-origin). Shells are typically defined at increasing radius intervals (e.g., 10Å, 20Å, 30Å, etc.).
- Shell-by-Shell Comparison: For each shell, calculate:
- Root Mean Square Deviation (RMSD) of rRNA atomic coordinates
- Sequence similarity of rRNA components
- Secondary structure conservation (percentage of A-form helices)
- Protein content and secondary structure composition
- Phylogenetic Mapping: Map conservation metrics against distance from PT-origin to establish relative evolutionary ages.
- Statistical Analysis: Perform correlation analysis between distance from PT-origin and conservation metrics to validate chronological significance.
Validation: The method is validated by consistent patterns observed across multiple phylogenetic lineages and by congruence with independent molecular clock analyses [15] [17].
Protocol: Assessing Proto-Ribosome Emergence Probability
Objective: To evaluate the statistical likelihood of spontaneous emergence of functional proto-ribosomes from random RNA pools.
Materials:
- Sequence and structural data for proposed proto-ribosome models
- Computational resources for probabilistic modeling
- Knowledge of RNA folding thermodynamics and dimerization energetics
Methodology:
- Constraint Identification: For each proto-ribosome model, identify the minimal set of nucleotide constraints necessary to preserve:
- Functional components (e.g., substrate binding sites)
- Tertiary structure integrity
- Dimerization interfaces (e.g., GNRA motifs)
- Probability Calculation: Compute the probability of random occurrence using:
- Monomer length and constrained position requirements
- "Limited specificity" considerations where appropriate
- Statistical mechanics of RNA folding and dimerization
- Energetic Assessment: Apply quantum mechanics computations to evaluate the stabilizing effect of dimerization for each model.
- Feasibility Threshold: Compare probabilities against prebiotically plausible RNA quantities (estimated based on early Earth conditions).
Interpretation: Models with probabilities requiring more than 1-10 kg of random RNA for statistically likely emergence are considered implausible for spontaneous prebiotic formation [14].
Diagram: Proto-Ribosome Feasibility Assessment Workflow - Methodological framework for evaluating spontaneous emergence likelihood of proto-ribosome models.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Ribosomal Evolutionary Studies
| Reagent/Category | Function/Application | Representative Examples |
|---|---|---|
| Ribosome Structures | Structural analysis and comparison | H. marismortui 50S (PDB: 1FFK), T. thermophilus 50S (PDB: 1GIY), E. coli ribosome structures |
| Structural Analysis Software | Molecular visualization, superposition, and measurement | PyMOL, Chimera, UCSF ChimeraX, Coot |
| Sequence Alignment Tools | Phylogenetic analysis and conservation mapping | ClustalOmega, MUSCLE, T-Coffee, RNA-specific aligners |
| Computational Chemistry Packages | Energetic calculations and molecular modeling | GROMACS, AMBER, Rosetta, Quantum chemistry packages |
| RNA Synthesis Systems | Experimental testing of ribozyme activity | In vitro transcription systems, Synthetic RNA oligonucleotides |
| Ribozyme Assay Components | Functional characterization of catalytic RNA | Radiolabeled nucleotides, Fluorescent tags, Substrate analogs |
The ribosome stands as a remarkable molecular fossil that preserves evidence of its evolutionary history within its contemporary architecture. Structural analyses confirm that the ribosome originated as an RNA machine that catalyzed peptide bond formation in the prebiotic RNA world. The "peeling the onion" methodology reveals a clear evolutionary chronology, with the most ancient components concentrated around the peptidyl transferase center and more recent additions located peripherally. Probabilistic assessments indicate that the dimeric proto-ribosome represents the most plausible starting point for the evolution of translation, as it is the only model with a realistic statistical likelihood of spontaneous emergence from random RNA polymers.
For drug development professionals, understanding the ribosome as an ancient RNA machine has practical implications. The functional core of the ribosome remains predominantly RNA-based, making it an attractive target for antibiotics that specifically interact with RNA structures. Many clinically important antibiotics (e.g., macrolides, tetracyclines, aminoglycosides) target ribosomal RNA, exploiting conserved features that trace back to ancient evolutionary origins. Furthermore, engineering novel ribozymes inspired by proto-ribosome principles holds promise for developing RNA-based therapeutics and synthetic biology applications [12].
Future research directions include experimental reconstruction of proposed proto-ribosome models, further exploration of the peptide-RNA partnerships that preceded the modern ribosome, and computational simulations of ribosome evolution. As structural biology techniques continue to advance, particularly in cryo-electron microscopy and molecular dynamics simulations, our ability to extract increasingly detailed evolutionary history from this molecular fossil will continue to grow, offering deeper insights into the origin of life and the transition from the RNA world to modern biological systems.
Ribonucleic acid (RNA) stands as a unique biopolymer capable of both storing genetic information and catalyzing biochemical reactions. This dual functionality is the cornerstone of the RNA World Hypothesis, a foundational concept in origins-of-life research which posits that early life forms were based on RNA prior to the evolutionary emergence of deoxyribonucleic acid (DNA) and proteins. This whitepaper provides a technical examination of RNA's roles, exploring the catalytic mechanisms of ribozymes, experimental demonstrations of prebiotic RNA reproduction, and the enduring fingerprints of the RNA world in modern biological systems. We further synthesize key quantitative data from foundational studies and outline essential methodological protocols, providing a resource for researchers exploring RNA biology and its applications in therapeutic development.
The RNA World Hypothesis represents a dominant paradigm for understanding the origin of life, proposing that during the primitive stages of life, RNA served as the primary genetic blueprint and catalytic workhorse [18]. This concept, first advanced in the 1960s by Carl Woese, Francis Crick, and Leslie Orgel, and later termed the "RNA World" by Walter Gilbert in 1986, resolves a fundamental paradox in life's origins: the interdependence of DNA, which requires proteins for replication, and proteins, which require DNA for specification [4] [3]. RNA elegantly bridges this gap with its dual capabilities. The hypothesis suggests that around 4 billion years ago, RNA-based life forms managed fundamental processes like replication and metabolism without the need for DNA or proteins, a period that paved the way for the eventual transition to the DNA-protein world observed in contemporary biology [18] [4].
RNA’s Structural Versatility and Functional Capacity
RNA's capacity for dual functionality is intrinsically linked to its chemical structure. While both RNA and DNA are nucleic acids, key distinctions endow RNA with greater functional versatility, albeit at the cost of stability.
Key Structural Distinctions from DNA
- Sugar Composition: RNA's backbone contains ribose sugar, which features a reactive hydroxyl group (-OH) on the 2' carbon. In contrast, DNA uses deoxyribose, which lacks this group, making DNA more resistant to hydrolysis and thus a more stable long-term information repository [18].
- Base Pairings: RNA utilizes uracil (U) instead of thymine (T) to pair with adenine (A) during complementary base pairing [18].
- Strandedness and Conformation: RNA is typically single-stranded, allowing it to fold into complex three-dimensional shapes through internal base pairing. This structural plasticity enables RNA to form catalytic pockets and interaction surfaces, a prerequisite for its enzymatic function [18]. DNA, with its consistent double-helical structure, is more rigid and serves primarily as a stable information storage medium.
Table 1: Comparative Structural and Functional Properties of RNA and DNA
| Property | RNA | DNA |
|---|---|---|
| Sugar Backbone | Ribose (with 2'-OH) | Deoxyribose (without 2'-OH) |
| Strandedness | Typically single-stranded | Typically double-stranded |
| Key Bases | Adenine (A), Uracil (U), Guanine (G), Cytosine (C) | Adenine (A), Thymine (T), Guanine (G), Cytosine (C) |
| Structural Conformation | Diverse secondary/tertiary structures | Uniform double-helix structure |
| Primary Function | Information transfer & catalysis | Stable genetic information storage |
| Chemical Stability | Lower (susceptible to hydrolysis) | Higher |
Catalytic RNA: The Ribozyme
The discovery of catalytic RNA, or ribozymes, provided the first definitive experimental evidence supporting the RNA World Hypothesis. Before this, it was believed that all biological catalysis was carried out by proteins (enzymes). The Nobel Prize-winning work of Sidney Altman and Thomas Cech in 1989 revealed that RNA segments could accelerate specific chemical reactions without being permanently altered, thus possessing enzymatic properties [4] [3].
Mechanisms and Modern Examples
Ribozymes catalyze a variety of reactions, including self-splicing of introns and cleavage of RNA strands [18]. Among the most well-studied are the hammerhead ribozymes, which are small, self-cleaving motifs found in plant viruses and other organisms [19]. These ribozymes fold into a characteristic structure that allows them to perform site-specific cleavage of their own phosphodiester backbone.
The most compelling modern example of a ribozyme is the ribosome. Despite being composed of both RNA and protein, the catalytic activity for peptide bond formation—the central reaction of protein synthesis—is performed by the ribosomal RNA (rRNA) component. This confirms that RNA is capable of catalyzing essential biosynthetic reactions and is a likely molecular fossil from the RNA world [4].
Experimental Models of Prebiotic RNA Replication
A significant challenge for the RNA World Hypothesis is explaining how the first RNA molecules could replicate without the assistance of modern protein enzymes. Research in prebiotic chemistry has made strides in demonstrating feasible non-enzymatic replication pathways.
Azoarcus Ribozyme Self-Assembly Model
Advanced experimental systems have been developed to study prebiotic RNA replication dynamics. One such model uses the Azoarcus tRNAIle intron, a ~200-nucleotide ribozyme that can be broken into multiple fragments (e.g., WXY and Z) [20]. These fragments can spontaneously reassemble into a covalently contiguous, functional ribozyme through a recombination reaction when incubated under appropriate conditions (e.g., 48°C with MgCl₂) [20].
This system allows researchers to create different RNA "genotypes" by altering short nucleotide sequences that govern fragment interaction specificity. By mixing these genotypes and tracking their reproduction over multiple serial dilution transfers, scientists can model frequency-dependent competition and cooperation among early RNAs, observing phenomena like stable coexistence in a "rock-paper-scissors" dynamic [20].
Table 2: Key Reagents for Prebiotic RNA Replication Studies (Azoarcus System)
| Research Reagent | Function/Description |
|---|---|
| Azoarcus Ribozyme Fragments (WXY, Z) | Core RNA building blocks designed to covalently self-assemble into a full-length, functional ribozyme. |
| MgCl₂ Buffer Solution | Provides essential divalent cations (Mg²⁺) that stabilize RNA tertiary structure and catalyze the transesterification assembly reaction. |
| Differentially Labeled Nucleotides (e.g., ³²P) | Allows for precise tracking and quantification of the reproduction rates of different RNA genotypes in a mixed competition experiment. |
| Serial Dilution Apparatus | Enables long-term evolution experiments by periodically transferring a fraction of the reaction to a fresh environment with new resources, mimicking natural selection. |
The following diagram illustrates the logical workflow and interactions within this experimental system:
Quantitative Dynamics of Prebiotic RNA Networks
Chemical game theory provides a quantitative framework for analyzing the interactions within networks of reproducing RNA molecules. In uncompartmentalized prebiotic scenarios, different RNA genotypes would have competed for common resources, such as nucleotide precursors or assembly factors.
Game Theory Analysis of RNA Interactions
Experiments with the Azoarcus ribozyme system quantify these interactions using a 2x2 payoff matrix [20]. The matrix elements represent the "payoff" or replication rate constant (e.g., the autocatalytic rate constant, kₐ) for one genotype when interacting with another. These values are derived from direct measurement of RNA assembly over time, often using differentially radiolabeled strands to track individual genotypes.
Table 3: Example Payoff Matrix for Two Interacting RNA Genotypes [20]
| Focal Genotype vs. Competing Genotype | Self-Assembly Rate Constant (kₐ) | Cross-Assembly Rate Constant (kₐ) |
|---|---|---|
| Genotype A vs. Genotype A | High (e.g., 0.45 min⁻¹) | Not Applicable |
| Genotype A vs. Genotype B | Reduced (e.g., 0.30 min⁻¹) | Measured value (e.g., 0.15 min⁻¹) |
| Genotype B vs. Genotype B | Low (e.g., 0.10 min⁻¹) | Not Applicable |
| Genotype B vs. Genotype A | Reduced (e.g., 0.08 min⁻¹) | Measured value (e.g., 0.25 min⁻¹) |
This data reveals whether the relationship between two genotypes is selfish, cooperative, or parasitic. For instance, a genotype might reproduce poorly in isolation but thrive in the presence of a partner that provides a catalytic benefit (molecular cooperation). These dynamics can predict stable equilibria in genotype frequencies or the emergence of multi-genotype ecosystems, such as the rock-paper-scissors analog observed with three RNA types [20].
"Molecular Fossils": Evidence in Contemporary Biology
The legacy of the RNA world is embedded in the core biochemistry of modern cells. Key cellular components and processes point to an ancient world dominated by RNA.
- The Ribosome: As noted, the ribosome is a ribozyme, with rRNA catalyzing peptide bond formation [4]. This is the strongest evidence for the RNA world, as it demonstrates RNA's capability to catalyze a central biological process.
- Other Non-Coding RNAs: Transfer RNA (tRNA) and messenger RNA (mRNA) are essential for protein synthesis. Furthermore, modern cells contain a variety of regulatory RNAs (e.g., miRNAs, siRNAs) and self-splicing introns that continue to perform catalytic and informational roles [18] [19].
- Coenzymes: Many essential metabolic coenzymes, such as acetyl-CoA and NADH, are ribonucleotide derivatives, suggesting they are molecular relics of an era where RNA was central to metabolism [18].
- Viroid-like Elements: Infectious circular RNAs like viroids and the hammerhead ribozymes found in modern RNA viruses are considered "living fossils." They exhibit key RNA world characteristics: they are small, non-coding, self-replicating, and catalytic [19]. Recent discoveries show these ribozymes are widespread in fungal and plant viruses, where some have been exapted for new roles, such as facilitating cap-independent translation initiation by functioning as an Internal Ribosome Entry Site (IRES) [19].
Challenges and Alternative Pathways
Despite its explanatory power, the RNA World Hypothesis faces significant challenges that drive ongoing research in prebiotic chemistry.
- Prebiotic Synthesis: The spontaneous formation of complex RNA molecules from a "primordial soup" of simple organic compounds remains difficult to explain. The specific conditions and pathways for forming the first nucleotides and polymerizing them into functional RNA are active areas of investigation [18] [11].
- Chemical Instability: RNA's relative instability, particularly the susceptibility of its ribose backbone to hydrolysis, poses a problem for its persistence in the harsh conditions of early Earth [4].
- Limited Catalytic Range: While ribozymes are versatile, they are generally not as efficient or diverse as protein enzymes in catalyzing the vast array of reactions required by modern cells [4].
These challenges have prompted research into even simpler genetic systems that might have preceded RNA, as well as scenarios where RNA and DNA emerged in parallel. For example, recent prebiotic chemistry research has identified a direct, non-enzymatic pathway for synthesizing DNA nucleosides from simple organic precursors, suggesting DNA subunits could have appeared alongside RNA much earlier than previously assumed [21].
RNA's dual capacity for genetic storage and catalysis provides a compelling solution to the puzzle of life's origins. The RNA World Hypothesis, supported by the discovery of ribozymes, the catalytic nature of the ribosome, and experimental models of RNA replication, offers a robust framework for understanding how life could have emerged from a prebiotic chemical environment. While challenges regarding the prebiotic synthesis of RNA persist, they serve as productive guides for future research.
The implications of this research extend beyond origins-of-life studies into biotechnology and medicine. Understanding ribozyme mechanics informs the design of synthetic ribozymes and aptamers for therapeutic and diagnostic applications. Furthermore, the principles of RNA-based catalysis and replication continue to inspire novel drug development strategies, including RNA-targeting therapies and the use of RNA in synthetic biology to create new functional systems. Continued interdisciplinary research bridging prebiotic chemistry, molecular biology, and biophysics will undoubtedly uncover deeper insights into RNA's foundational role in the story of life.
The origin of life presents a fundamental "chicken-and-egg" conundrum that has long challenged researchers. In modern biological systems, nucleic acids (RNA and DNA) store and transmit genetic information, while proteins execute most catalytic functions. However, assembling proteins requires the information encoded in nucleic acids, while synthesizing and replicating nucleic acids themselves typically requires protein enzymes [22]. Similarly, protein enzymes drive metabolism, yet their components owe their existence to metabolic processes [22]. This reciprocal dependency creates an apparent paradox for understanding how life could emerge from prebiotic chemistry.
The RNA world hypothesis provides a compelling solution to this dilemma by proposing that early life forms utilized RNA molecules that served both genetic and catalytic functions [23]. This concept suggests that RNA alone could have been both the "chicken and the egg"—capable of storing information and catalyzing the chemical reactions necessary for self-replication [8] [24]. The discovery of ribozymes (RNA molecules with enzymatic activity) provided critical experimental support for this hypothesis, demonstrating that RNA can indeed perform both these essential functions [22] [3]. This framework transforms our understanding of life's origins, suggesting a plausible pathway from prebiotic chemistry to simple biological systems through the intermediary of self-replicating RNA molecules housed within primitive protocells.
The RNA World: A Theoretical Framework
Historical Development and Key Principles
The conceptual foundation for the RNA world hypothesis was established in the 1960s through the independent work of several prominent scientists. Francis Crick, Leslie Orgel, and Carl Woese first suggested that RNA might have played a more central role in early life forms [3]. Critical insights emerged when Robert W. Holley determined the sequence and structure of transfer RNA (tRNA), revealing intricate folds that resembled proteins more than the simple double helix of DNA [22]. Orgel and Crick immediately recognized the implication: RNA might have performed protein-like enzymatic functions during life's origin.
The hypothesis gained significant traction in the early 1980s when Sidney Altman and Thomas Cech made the groundbreaking discovery that RNA can indeed act as an enzyme—catalyzing specific chemical reactions [22]. This discovery of ribozymes earned them the Nobel Prize in Chemistry in 1989 and provided the first experimental evidence that RNA could potentially catalyze its own replication [3]. The term "RNA world" was subsequently coined by Walter Gilbert in 1986, encapsulating the concept of an early evolutionary stage dominated by multifunctional RNA molecules [22] [3].
The RNA world hypothesis posits several key principles:
- RNA preceded DNA and proteins as the primary biological macromolecule [23]
- Early RNA molecules served both as repositories of genetic information and as catalysts for metabolic reactions [24]
- Self-replicating RNA systems constituted the first simple life forms [22]
- DNA and proteins emerged later through RNA-based evolution, eventually assuming more specialized roles due to their superior stability and catalytic efficiency [3]
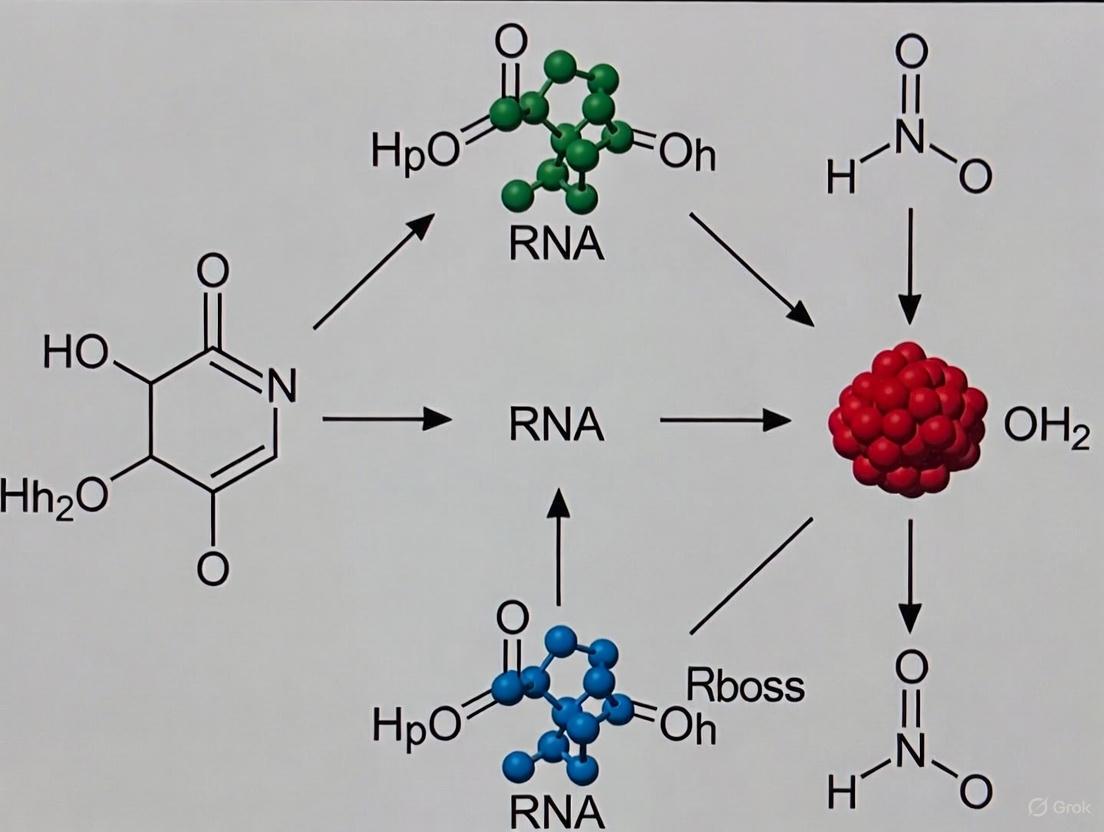
Mechanistic Workflow of RNA-Centric Early Evolution
The following diagram illustrates the proposed stepwise process through which RNA-based life could have emerged from prebiotic chemistry and eventually given rise to modern biological systems:
This evolutionary sequence represents a progression from simple chemistry to increasingly complex biological systems, with RNA serving as the central player throughout the early stages. The transition from the prebiotic world to the RNA world represents the most critical phase, where natural processes first gave rise to self-sustaining, evolving molecular systems [22] [23].
Experimental Evidence for Prebiotic RNA Replication
Key Experimental Methodologies
Research into prebiotic RNA replication has employed several sophisticated experimental approaches to simulate early Earth conditions and test specific aspects of the RNA world hypothesis. The table below summarizes core methodologies used in this field:
Table 1: Key Experimental Methods in Prebiotic RNA Replication Research
| Method | Experimental Approach | Key Insights Generated |
|---|---|---|
| Prebiotic Synthesis Simulations | Recreating early Earth conditions (temperature, pH, mineral surfaces) to test RNA component formation [24] | Demonstrated plausible pathways for pyrimidine nucleotide synthesis; identified challenges in ribose stability [24] |
| Ribozyme Engineering | In vitro selection (SELEX) to identify RNA sequences with catalytic functions [24] | Discovered ribozymes capable of ligation, replication, and metabolic functions; revealed catalytic potential of even small RNAs [22] |
| Protocell Models | Constructing primitive membrane-bound compartments from fatty acids [22] | Showed how primitive cells could grow, divide, and retain genetic material without complex protein machinery [22] |
| Phosphorylation Studies | Testing phosphorylation efficiency of different sugars under prebiotic conditions [25] | Revealed ribose's selective advantage in phosphorylation reactions critical for nucleotide formation [25] |
Critical Research Reagents and Solutions
The experimental investigation of prebiotic RNA replication requires specialized reagents that mimic proposed early Earth conditions. The following table details essential research reagents and their functions in this field:
Table 2: Essential Research Reagents for Prebiotic RNA Replication Studies
| Reagent | Composition/Type | Function in Experiments |
|---|---|---|
| Diamidophosphate (DAP) | Phosphorus-containing compound | Serves as prebiotic phosphate donor in phosphorylation reactions; enables ribose phosphorylation without enzyme catalysis [25] |
| Ribose and Alternative Sugars | Pentose sugars (ribose, arabinose, lyxose, xylose) | Comparative substrates for testing phosphorylation selectivity; determine ribose's potential competitive advantages [25] |
| Fatty Acid Vesicles | Membranous structures composed of fatty acids | Model primitive protocells; study compartmentalization effects on RNA replication and selection [22] |
| Clay Minerals | Montmorillonite, kaolinite, etc. | Provide catalytic surfaces for RNA assembly and organization; enhance RNA oligomerization under prebiotic conditions [23] |
| Short RNA Oligomers | Synthetic RNA sequences of varying lengths | Substrates for testing ribozyme activity, replication fidelity, and template-directed synthesis [24] |
Experimental Workflow for Ribose Selectivity Studies
Recent investigations into why ribose became the sugar of choice for RNA have followed a systematic experimental approach. The following diagram outlines the methodology used in groundbreaking phosphorylation studies:
This methodology revealed that ribose phosphorylates more rapidly and selectively than other similar sugars when exposed to diamidophosphate under prebiotic conditions [25]. Specifically, the research demonstrated that ribose phosphorylation occurred at a much faster rate and exclusively produced the five-member ring structure found in contemporary RNA, while other sugars formed mixtures of five- and six-member rings [25]. This selective advantage may have been a critical factor in ribose emerging as the sugar component of early genetic molecules.
Current Challenges and Research Frontiers
Key Limitations of the RNA World Hypothesis
Despite its explanatory power and experimental support, the RNA world hypothesis faces several significant challenges that represent active research frontiers:
Prebiotic Synthesis Difficulties: The spontaneous formation of RNA nucleotides under plausible early Earth conditions remains chemically challenging. While recent work has identified potential pathways for pyrimidine nucleotide synthesis, significant hurdles remain in explaining the prebiotic formation of purine nucleotides and the selection of specific sugar configurations [24].
Chemical Instability: RNA is inherently less stable than DNA, particularly at moderate temperatures and alkaline pH. The RNA backbone is susceptible to hydrolysis, especially in the presence of divalent metal ions like Mg²⁺ that are also important for RNA folding and function [24]. Research suggests this limitation might have been mitigated by acidic environmental conditions or freezing temperatures [24].
Limited Catalytic Repertoire: While ribozymes can catalyze various reactions, their catalytic efficiency and diversity generally fall short of protein enzymes. This has led to questions about whether RNA alone could have catalyzed the full range of reactions necessary for primitive metabolism [8] [24].
Template-Directed Replication: Establishing a robust system of non-enzymatic RNA replication remains a significant challenge. While short RNA sequences can template complementary strand formation, achieving accurate and efficient copying of longer functional RNA sequences without modern enzymatic machinery has proven difficult [22].
Quantitative Data on RNA Stability and Reactivity
Research has generated important quantitative data relevant to the stability and reactivity of RNA under various prebiotic conditions:
Table 3: Experimental Data on RNA Stability and Reactivity Under Prebiotic Conditions
| Parameter | Experimental Conditions | Key Findings | Research Implications |
|---|---|---|---|
| Optimal Ribozyme Activity | -7°C to -8°C in eutectic ice phases [24] | Maximum ribozyme activity observed at subzero temperatures | Suggests cold environments may have protected and enhanced early RNA function [24] |
| pH Stability | Acidic conditions (pH 4-5) [24] | Enhanced phosphodiester bond and aminoacyl ester bond stability | Supports potential for RNA world evolution in acidic environments [24] |
| Ribose Phosphorylation Rate | Comparative phosphorylation of four pentose sugars with DAP [25] | Ribose phosphorylated significantly faster than arabinose, lyxose, or xylose | Demonstrates selective chemical advantage for ribose incorporation into early nucleotides [25] |
| Backbone Heterogeneity Tolerance | RNA with mixed 2'-5' and 3'-5' linkages [24] | Partial ribozyme function retained with non-standard backbone configurations | Suggests early RNA systems may have been more structurally flexible than modern RNA [24] |
The RNA world hypothesis, while not without challenges, remains the most compelling framework for understanding how life emerged from prebiotic chemistry. By positing that RNA molecules served both genetic and catalytic functions in early evolution, this hypothesis elegantly resolves the fundamental "chicken-and-egg" dilemma of life's origin. Experimental evidence continues to accumulate, from the discovery of catalytic RNA to recent advances in understanding the selective chemical advantages of ribose phosphorylation.
Ongoing research focuses on addressing the remaining challenges, particularly regarding prebiotic nucleotide synthesis, RNA stability, and the mechanisms of early RNA replication. As investigation continues across multiple disciplines—including chemistry, biology, and planetary science—our understanding of this critical transition from non-living chemistry to biological systems continues to deepen. The solution to the prebiotic RNA replication problem not only illuminates life's origins on Earth but also informs the search for life elsewhere in the universe by identifying potential universal principles of biogenesis.
A series of landmark studies published in 2025 has demonstrated for the first time how amino acids could spontaneously attach to RNA under plausible early Earth conditions, providing a long-sought mechanistic bridge between genetics and metabolism in origin-of-life research. This whitepaper details the experimental protocols, quantitative findings, and significant implications of these discoveries, which directly address fundamental challenges within the RNA World Hypothesis. We present comprehensive data tabulation, experimental workflows, and analytical frameworks to contextualize these findings for researchers investigating prebiotic chemistry, molecular evolution, and the origins of biological information systems.
The RNA World Hypothesis represents a dominant theoretical framework for understanding the origin of life, proposing that self-replicating RNA molecules served as the precursor to all current life forms [4] [3]. First conceptualized by Carl Woese, Francis Crick, and Leslie Orgel in the 1960s and later termed by Walter Gilbert in 1986, this hypothesis posits that RNA once carried out both genetic information storage and catalytic functions independently before the evolutionary emergence of DNA and proteins [4]. The discovery of ribozymes (catalytic RNA molecules) by Sidney Altman and Thomas Cech provided critical support for this hypothesis, demonstrating RNA's capacity for enzymatic catalysis [4] [3].
Despite its theoretical appeal, the RNA World Hypothesis faces significant challenges that have long constrained its acceptance as a complete model for abiogenesis. As noted in a 2012 critical review, these objections include: (i) the prebiotic implausibility of RNA's complex molecular structure, (ii) RNA's inherent chemical instability, (iii) the relative rarity of catalytic activity among RNA sequences, and (iv) RNA's limited catalytic repertoire compared to proteins [8]. NASA's 1996 report similarly highlighted the "significant difficulties" surrounding RNA's chemical fragility and narrow catalytic range [4].
The most persistent paradox has been the chicken-and-egg problem of molecular interdependence: modern biology requires proteins to synthesize nucleic acids and nucleic acids to synthesize proteins [26]. Until recently, no plausible prebiotic mechanism existed to explain how RNA and amino acids initially established their functional relationship, creating a fundamental gap in our understanding of how the RNA world evolved toward the peptide/RNA world and eventually to DNA-based life [27].
Recent Breakthroughs: Bridging the RNA-Amino Acid Divide
Thioester-Mediated RNA Aminoacylation (Powner Lab, UCL)
A landmark study published in Nature in August 2025 by researchers at University College London demonstrated a spontaneous chemical mechanism for linking amino acids to RNA [28] [26]. The team showed that thioesters – high-energy organic sulfur compounds derived from pantetheine (the active core of coenzyme A) – could facilitate direct amino acid attachment to RNA strands in water at neutral pH without enzymatic catalysis [28].
Table 1: Key Experimental Findings from Thioester-Mediated Aminoacylation Study
| Experimental Parameter | Specific Conditions | Observed Outcome |
|---|---|---|
| Reaction Environment | Aqueous solution, neutral pH | Successful aminoacylation without organic solvents or extreme conditions |
| Temperature Range | Room temperature to just above freezing | Robust reaction across varied prebiotically plausible temperatures |
| Chemical Activator | Aminoacyl-thiols (thioesters) derived from pantetheine | Selective RNA modification over competing side reactions |
| Site Specificity | 2',3'-diol position on RNA ribose | Same site used in modern biological aminoacylation |
| Amino Acids Tested | Arginine, glycine, alanine | Successful attachment for multiple proteinogenic amino acids |
| Additional Discovery | Eutectic phases (ice formation) | Enhanced reaction efficiency through solute concentration |
This research successfully united elements of two competing origin-of-life theories: the "RNA world" (emphasizing primordial genetic molecules) and the "thioester world" (prioritizing early metabolic energy cycles) [28] [26]. The demonstrated chemistry is particularly significant because it proceeds without the need for highly reactive molecules that break down in water – a limitation that had thwarted previous attempts since the 1970s [28].
Autocatalytic Chimeric Ribozymes (Szostak Lab)
Complementary research from Jack Szostak's lab published in Science Advances in 2025 described a self-replicating chimeric ribozyme composed of RNA strands bridged by intervening amino acids [29]. This hybrid molecule demonstrated dual functionality: the capacity to make more of itself from smaller fragments and the ability to build other chimeric amino acid-bridged RNA molecules.
The researchers constructed a detailed computational kinetic model that confirmed the observed behavior could only be explained by autocatalytic kinetics, with the reaction rate increasing as products accelerated further production [29]. The process functioned across a wide range of temperatures (from just above freezing to room temperature) and varying acidity levels, indicating resilience in diverse early Earth environments [29].
Table 2: Properties of Autocatalytic Chimeric Ribozymes
| Property | Characterization | Prebiotic Significance |
|---|---|---|
| Catalytic Function | Self-replication and assembly of unrelated chimeric ribozymes | Suggests capacity for molecular evolution and diversification |
| Structural Composition | Long RNA pieces bridged by intervening amino acids | Amino acids act as "molecular glue" in assembly |
| Kinetic Behavior | Autocatalytic, with verified self-replicating kinetics | Explains emergence of self-sustaining chemical systems |
| Environmental Robustness | Functions across wide temperature and pH ranges | Plausible in diverse early Earth environments |
| Evolutionary Potential | Can synthesize functionally unrelated chimeric ribozymes | Acts as general "assembler" in primordial molecular machinery |
Amino Acid Catalysis of RNA Formation
A June 2025 study in Nature Communications revealed that amino acids can significantly catalyze RNA formation under ambient alkaline conditions, demonstrating a reciprocal relationship between these fundamental biomolecules [30]. Researchers found that amino acids, without additional chemical activators, promoted RNA copolymerization more than 100-fold starting from prebiotically plausible ribonucleoside-2',3'-cyclic phosphates (cNMPs) [30].
The catalytic effect was explained by acid-base catalysis, with optimal efficiency at pH values near the amine pKaH. The fold-change in oligomerization yield was nucleobase-selective, resulting in increased compositional diversity necessary for subsequent molecular evolution and favoring the formation of natural 3'−5' linkages [30]. This discovery reveals a clear functional role for amino acids in RNA evolution earlier than previously assumed and helps explain how sufficient RNA diversity and length emerged to initiate self-replication cycles.
Experimental Protocols and Methodologies
Thioester-Mediated Aminoacylation Protocol
The UCL team employed a biochemically-inspired approach using thioesters to activate amino acids for RNA attachment [28] [26]. The specific methodology included:
1. Thioester Formation:
- Amino acids were reacted with pantetheine to form aminoacyl-thiols (thioesters)
- Pantetheine was selected due to its prebiotic plausibility and presence as the active core of coenzyme A in all living cells
- This activation step converted the amino acids into a more reactive form while maintaining stability in aqueous environments
2. Reaction Conditions:
- Reactions were conducted in pure water at neutral pH (approximately 7.0)
- No enzymes, ribosomes, or cellular components were required
- Temperature variations were tested from near-freezing to room temperature
- In some experiments, freezing was utilized to create eutectic phases that concentrated reactants
3. Analytical Techniques:
- Multiple magnetic resonance imaging (MRI) techniques to determine atomic arrangement
- Mass spectrometry to determine molecular sizes
- High-performance liquid chromatography (HPLC) for separation and quantification
- The absence of extreme heat, volcanic vents, or complex pre-activation distinguished this protocol from previous attempts
Autocatalytic Ribozyme Assembly Protocol
The Szostak lab's approach focused on demonstrating and verifying self-replicating molecular systems [29]:
1. Molecular Design:
- Construction of chimeric ribozymes combining RNA components with bridging amino acids
- Design of fragment molecules that could serve as building blocks for self-assembly
2. Kinetic Analysis:
- Careful measurement of each reaction within the complex system
- Development of a computational kinetic model to verify autocatalytic behavior
- Elimination of alternative explanations such as chance resemblance to self-replicating kinetics
3. Environmental Testing:
- Evaluation of function across temperature gradients (0°C to 25°C)
- Assessment of performance under varying acidity levels
- Demonstration of robustness in diverse plausible prebiotic conditions
Experimental Workflow Visualization
The following diagram illustrates the key experimental workflow for the thioester-mediated RNA aminoacylation demonstrated in the 2025 Nature study:
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Research Reagents for Prebiotic RNA-Amino Acid Studies
| Reagent / Material | Function in Experimental System | Prebiotic Plausibility |
|---|---|---|
| Pantetheine | Forms thioester intermediates with amino acids; serves as molecular activator | Demonstrated prebiotic synthesis; core of universal coenzyme A |
| Ribonucleoside-2',3'-cyclic phosphates (cNMPs) | RNA building blocks; spontaneously oligomerize under appropriate conditions | Products of prebiotic nucleotide synthesis and RNA cleavage |
| Aminoacyl-thiols | Activated amino acid derivatives; enable selective RNA modification without enzymes | Formed from amino acids and thiol compounds in prebiotically plausible conditions |
| Chimeric ribozyme constructs | Demonstrate self-replication and catalytic assembly of molecular hybrids | Model potential transitional forms between RNA and peptide/RNA worlds |
| Hydrophobic amino acids (Val, Leu, Ile) | Catalyze RNA oligomerization from cNMPs under alkaline conditions | Available from prebiotic synthesis and meteoritic delivery |
Biochemical Pathway and Evolutionary Significance
The spontaneous connection between RNA and amino acids represents a crucial transitional step in early molecular evolution. The following diagram illustrates the biochemical pathway and its significance in bridging prebiotic chemistry toward biological systems:
This biochemical pathway demonstrates how simple prebiotic chemistry could have transitioned toward biological complexity through the following evolutionary stages:
- Prebiotic Chemistry: Random assembly of monomers in early Earth environments [27]
- Molecular Activation: Thioester-mediated activation provides energy for bond formation [28]
- Stable Linkage: Covalent RNA-amino acid complexes create selectable functional units [31]
- Primitive Peptide Synthesis: Short peptides form without complex machinery [28]
- Molecular Selection: Functional advantages drive evolutionary pressure [27]
- Code Emergence: Specificity in RNA-amino acid interactions lays foundation for genetic code [27]
Research Implications and Future Directions
Implications for Origin of Life Research
These findings fundamentally reshape our understanding of early molecular evolution by:
- Resolving Key Paradoxes: The chicken-and-egg problem of molecular interdependence is addressed through demonstration of spontaneous, non-enzymatic linkage [26]
- Unifying Theoretical Frameworks: The research bridges the "RNA world" and "metabolism-first" hypotheses, suggesting co-evolution rather than sequential emergence [28]
- Providing Evolutionary Pathways: The demonstrated chemistry offers plausible steps from prebiotic chemistry to the peptide/RNA world [27]
- Explaining Biochemical Universals: The privileged role of adenosine monophosphate (AMP) in many pivotal biomolecules (ATP, NAD+, FAD, CoA) may reflect its high reactivity in these primordial connection chemistries [31]
Technical Applications and Research Opportunities
For researchers and drug development professionals, these discoveries open several promising avenues:
- Abiogenic Synthesis Approaches: Development of novel biomolecule synthesis methods inspired by prebiotic chemistry
- Ribozyme Engineering: Design of improved catalytic nucleic acids using amino acid cofactors
- Drug Discovery Platforms: Utilization of chimeric amino acid-RNA molecules as novel therapeutic targets
- Origin-of-Life Simulation: Enhanced computational models incorporating these connection mechanisms
Unanswered Questions and Research Challenges
Despite these significant advances, important questions remain:
- Sequence Specificity: How did RNA sequences evolve to bind preferentially to specific amino acids, leading to the genetic code? [28]
- Environmental Constraints: Which specific early Earth environments most favored these reactions? [26]
- Evolutionary Transition: What molecular pathways connected these simple attachment chemistries to the sophisticated ribosomal machinery of modern biology? [27]
- Prebiotic Source: What were the predominant prebiotic sources of key components like pantetheine? [28]
Future research directions should focus on establishing how RNA sequences developed specificity for particular amino acids, investigating more complex prebiotic reaction networks, and exploring how these primitive systems transitioned toward modern biological machinery.
The recent demonstrations of spontaneous RNA-amino acid connections under plausible early Earth conditions represent a transformative advancement in origin-of-life research. By providing experimentally verified mechanisms for bridging the historical divide between genetics and metabolism, these findings address long-standing objections to the RNA World Hypothesis while suggesting a more integrated, co-evolutionary model for life's emergence.
The methodological approaches, quantitative data, and theoretical frameworks presented in this whitepaper provide researchers with both the technical foundations and conceptual tools to further explore these fundamental biochemical relationships. As the field progresses, these discoveries promise to illuminate not only life's ancient origins but also new approaches to biomolecular engineering and therapeutic development.
The origin of life represents one of science's most fundamental challenges, characterized by the apparent paradox of the interdependent relationship between nucleic acids and proteins. For decades, the "RNA world" and "thioester world" hypotheses have stood as competing models for prebiotic chemistry. This whitepaper examines a groundbreaking experimental advance that bridges these two frameworks, demonstrating through detailed chemical methodology how thioester-activated amino acids spontaneously aminoacylate RNA under plausible early-Earth conditions. We present comprehensive quantitative data, experimental protocols, and analytical frameworks that collectively support a unified model for the emergence of nucleotide-directed peptide biosynthesis, offering new perspectives for origins of life research and synthetic biology applications.
The quest to understand life's origins has long been dominated by two prominent yet seemingly contradictory hypotheses: the "RNA world" and the "thioester world." The RNA world hypothesis, first formally proposed by Walter Gilbert in 1986 but with roots reaching back to the 1960s, posits that self-replicating RNA molecules served as the initial hereditary and catalytic systems before the evolution of DNA and proteins [32]. This theory resolves the chicken-and-egg dilemma of molecular biology by proposing a single molecule capable of both information storage and catalysis. In contrast, the "thioester world" hypothesis, advanced by Nobel laureate Christian de Duve, suggests that energy-rich thioester compounds drove primitive metabolic cycles before the emergence of genetic systems [33].
The fundamental challenge lies in the interconnectedness of modern biology: proteins (encoded by nucleic acids) are required for nucleic acid replication, creating an evolutionary paradox. While the RNA world theory is supported by RNA's dual capabilities as both a genetic polymer and catalyst (ribozymes), it struggles to explain the origins of protein synthesis [32]. Conversely, the thioester world offers a plausible energy source for early chemistry but lacks a mechanism for heredity. A synthesis of these frameworks provides a more complete model for life's emergence, suggesting that thioester-driven metabolism and RNA-based information systems co-evolved, each enabling the other's sophistication.
Theoretical Foundations
The RNA World Hypothesis
The RNA world hypothesis rests on several key observations regarding RNA's unique biochemical properties. First, RNA can store genetic information through its sequence of nucleotides, analogous to DNA. Second, RNA possesses catalytic capabilities as demonstrated by ribozymes, which perform essential functions in modern cells, including protein synthesis in the ribosome [32]. The discovery that the catalytic core of the ribosome is composed of RNA, not protein, provides compelling evidence for RNA's primordial role. Furthermore, RNA plays central roles in fundamental biological processes: transfer RNA (tRNA) and messenger RNA (mRNA) in protein synthesis, and numerous regulatory RNAs in gene expression.
Theoretical work indicates that early RNA molecules could have performed a range of functions necessary for primitive life:
- Self-replication: Laboratory experiments have demonstrated RNA sequences capable of template-directed replication [32].
- Simple catalysis: Ribozymes can catalyze various chemical reactions including formation of peptide bonds [32].
- Metabolic cofactors: Many essential cofactors (e.g., NAD, CoA) contain adenosine moieties, suggesting ancient RNA-world "vestiges" [32].
However, the RNA world hypothesis faces significant challenges, particularly regarding prebiotic RNA synthesis and the origins of the RNA-protein relationship.
The Thioester World Hypothesis
Christian de Duve's thioester world hypothesis emphasizes metabolism-first origins, proposing that energy-rich thioester compounds provided the thermodynamic driving force for early chemical evolution. Thioesters, which feature a sulfur atom bonded to an acyl group, are high-energy compounds that play central roles in modern metabolism, including the citric acid cycle, fatty acid biosynthesis, and non-ribosomal peptide synthesis [33] [34].
De Duve argued that thioesters could have formed abiotically on early Earth and driven the polymerization of amino acids into peptides before the emergence of replicating systems. This framework addresses the energy problem that plagues many prebiotic scenarios—how endergonic reactions necessary for building biological molecules could proceed efficiently. The thioester world posits that simple thioester-driven peptide cycles gradually increased in complexity, eventually incorporating nucleic acids into a more sophisticated biochemical network.
Table 1: Key Characteristics of RNA World and Thioester World Hypotheses
| Feature | RNA World Hypothesis | Thioester World Hypothesis |
|---|---|---|
| Primary focus | Information storage and replication | Energy metabolism and catalysis |
| Key molecules | Ribonucleotides, RNA polymers | Thioesters, peptides, coenzyme A |
| Strengths | Explains genetic code origin; RNA has catalytic capacity | Solves energy problem; connects to core metabolism |
| Limitations | Prebiotic RNA synthesis challenging; protein synthesis origin unexplained | Lacks heredity mechanism; information transfer unclear |
| Modern evidence | Ribozymes, RNA in replication/translation apparatus | Ubiquity of thioesters in central metabolic pathways |
Experimental Breakthrough: Unified Model
A landmark study published in Nature in August 2025 by Singh et al. provides the first experimental demonstration bridging the RNA and thioester worlds [35]. The research team from University College London, led by Professor Matthew Powner, achieved spontaneous aminoacylation of RNA using thioester-activated amino acids under conditions plausible for early Earth—neutral pH water at room temperature [28] [36].
This work addresses what Powner describes as "the most intriguing causal paradox in biology"—the origin of protein synthesis, where proteins are required to make proteins [35]. The study demonstrates that aminoacyl-thiols (thioester-activated amino acids) react selectively with RNA diols over amine nucleophiles, promoting aminoacylation over non-coded peptide bond formation [35]. This selectivity had never been achieved previously in aqueous solution and represents a critical step toward understanding how RNA might have first come to control protein synthesis.
Core Chemical Methodology
The experimental protocol centers on using aminoacyl-thiols to achieve selective RNA aminoacylation. The researchers found that biological thioesters provide ideal activation energy—sufficient to drive the reaction but mild enough to maintain selectivity [35]. The methodology proceeds through several key stages:
- Amino acid activation: Proteinogenic amino acids were converted to aminoacyl-thiols using pantetheine, a sulfur-containing compound and functional fragment of coenzyme A [28] [35].
- RNA aminoacylation: The resulting aminoacyl-thiols were combined with RNA in aqueous solution at neutral pH, leading to spontaneous aminoacylation at the RNA's 2',3'-diol moiety [35].
- Peptide synthesis: By switching from thioester to thioacid activation, the researchers inverted diol/amine selectivity, promoting peptide bond formation with aminoacylated RNA [35].
Remarkably, the team demonstrated both RNA aminoacylation and subsequent peptide synthesis in a one-pot reaction without enzymes, purification steps, or template-directed reactivity [35] [37]. The process showed broad side-chain compatibility, working effectively with 14 different proteinogenic amino acids including Ala, Arg, Asp, Glu, Gly, His, Leu, Lys, Phe, Pro, Ser, and Val [35].
Diagram 1: Unified prebiotic chemistry workflow (16 words)
Quantitative Results and Analysis
The research provides comprehensive quantitative data supporting the efficiency and selectivity of thioester-mediated RNA aminoacylation. The reaction demonstrated remarkable chemoselectivity, with aminoacyl-thiols preferring RNA diols over competing amine nucleophiles by significant margins [35]. This selectivity is crucial as it prevents uncontrolled peptide synthesis, enabling RNA-directed protein formation.
Table 2: Aminoacylation Efficiency Across Selected Amino Acids
| Amino Acid | Relative Aminoacylation Efficiency | Key Observations |
|---|---|---|
| Alanine | High | Stable aminoacyl-RNA formation |
| Arginine | Enhanced | Unprecedented side-chain nucleophilic catalysis |
| Glycine | High | Efficient dipeptide formation |
| Histidine | Moderate | Compatible with imidazole side chain |
| Leucine | High | Representative of hydrophobic amino acids |
| Phenylalanine | High | Aromatic side chain compatible |
| Serine | Moderate | Hydroxyl group does not interfere |
The study further demonstrated that duplex formation directs chemoselective 2',3'-aminoacylation of RNA, mimicking the structural context of modern tRNA [35] [37]. When double-stranded RNA was used—more similar to actual tRNA structure—aminoacylation occurred preferentially at the 3' end, analogous to biological systems [37]. Environmental conditions such as freezing—which creates eutectic phases that concentrate reactants—were found to enhance the aminoacylation process [26].
Experimental Protocols
Thioester-Mediated RNA Aminoacylation
Principle: Aminoacyl-thiols (1) react selectively with RNA 2',3'-diols over amine nucleophiles in neutral pH water, enabling the formation of aminoacyl-RNA without enzymatic catalysis [35].
Materials:
- Proteinogenic amino acids (e.g., Ala, Arg, Gly, Leu)
- Pantetheine (or related thiol cofactors)
- RNA oligonucleotides (single-stranded and duplex)
- Aqueous buffer (pH 7.0, neither acidic nor alkaline)
- Standard laboratory glassware
Procedure:
- Amino acid activation: Prepare aminoacyl-thiols (1) by reacting amino acids with pantetheine in aqueous solution. The reaction proceeds spontaneously at room temperature [35].
- RNA aminoacylation: Combine aminoacyl-thiols with RNA oligonucleotides in neutral pH buffer. The optimal RNA concentration should be sufficient to facilitate molecular interactions (reactions in dilute ocean conditions are less effective) [28] [26].
- Incubation: Allow the reaction to proceed at ambient temperature (20-25°C) or under freezing conditions to enhance yield through eutectic concentration effects [26].
- Analysis: Monitor reaction progress using:
- Magnetic resonance imaging (to determine atomic arrangement)
- Mass spectrometry (to determine molecular size)
- Chromatographic techniques to separate and identify products [28]
Key Observations:
- Aminoacyl-thiols (1) exhibit remarkable stability in water, with hydrolysis dominating over peptide formation across all pH levels tested [35].
- Reaction selectivity favors RNA diols over amine nucleophiles, suppressing adventitious peptide bond formation [35].
- Double-stranded RNA directs aminoacylation to the 3' end, mimicking biological tRNA charging [37].
Peptidyl-RNA Synthesis
Principle: A switch from thioester to thioacid activation inverts diol/amine selectivity, promoting peptide bond formation with aminoacylated RNA [35].
Procedure:
- Prepare aminoacyl-RNA: First generate aminoacyl-RNA as described in Protocol 4.1.
- Add aminothioacid: Introduce aminothioacid molecules carrying additional amino acids to the reaction mixture.
- Oxidative activation: Include an oxidizing agent to facilitate peptide bond formation between the amino acid on the aminoacylated RNA and the new amino acid [37].
- One-pot reaction: Conduct both aminoacylation and peptide synthesis sequentially in the same reaction vessel without purification of intermediate aminoacyl-RNAs [37].
Key Observations:
- Two-step, one-pot formation of peptidyl-RNA occurs efficiently in water at neutral pH [35].
- The process requires no evolved catalysts, intramolecular reactivity, or Watson-Crick templated interactions [35].
- This method achieves high-yielding, chemoselective synthesis of peptidyl-RNA, representing a plausible prebiotic precursor to modern protein synthesis [35] [37].
Research Reagent Solutions
Table 3: Essential Research Reagents for Prebiotic Chemistry Studies
| Reagent | Function/Application | Prebiotic Relevance |
|---|---|---|
| Pantetheine | Sulfur-bearing compound for thioester formation; core of coenzyme A | Demonstrated synthesis under early Earth conditions; universal in modern metabolism [28] [33] |
| Aminoacyl-thiols | Activated amino acids for selective RNA aminoacylation | Bridge thioester and RNA worlds; enable RNA charging without enzymes [35] |
| RNA oligonucleotides | Substrate for aminoacylation; model for early genetic material | Single-stranded and duplex forms test structural effects on reactivity [37] |
| Aminothioacids | Peptide bond formation with aminoacyl-RNA | Enable extension to peptide synthesis under same conditions [35] |
| Neutral pH aqueous buffer | Reaction medium simulating early Earth water bodies | Plausible prebiotic environment; enables chemistry in "ponds or lakes" [28] [26] |
Implications and Future Research Directions
The unification of RNA world and thioester world hypotheses through demonstrated chemistry has profound implications for origins of life research and beyond. For the field of prebiotic chemistry, it provides a plausible pathway for the emergence of nucleotide-directed protein synthesis, addressing a fundamental paradox in life's origins. The experimental evidence that simple thioesters can mediate the specific attachment of amino acids to RNA under mild conditions suggests that the genetic code could have emerged from straightforward chemical principles rather than requiring complex pre-existing molecular machinery.
For synthetic biology and drug development, these findings offer new approaches for creating hybrid biomolecules and simplified protein synthesis systems. The demonstrated ability to form peptidyl-RNA conjugates without enzymes or the ribosome suggests novel strategies for synthesizing modified peptides or creating artificial translation systems. Pharmaceutical researchers might exploit similar chemistry to develop new RNA-peptide hybrid therapeutics or simplified in vitro evolution systems.
Future research directions emerging from this work include:
- Sequence specificity: Determining how specific RNA sequences could develop preferential binding to particular amino acids—the origin of the genetic code [28] [37].
- Protocellular integration: Exploring how these processes function within membrane-bound compartments or on mineral surfaces.
- Extended chemistry: Investigating whether similar principles apply to other biological polymers and cofactors.
- Astrobiological applications: Applying these insights to the search for life elsewhere in our solar system, particularly in sulfur-rich environments like those found on Europa or Enceladus [26].
As Powner notes, "There are numerous problems to overcome before we can fully elucidate the origin of life, but the most challenging and exciting remains the origins of protein synthesis" [28]. This research provides a robust chemical foundation upon which to build increasingly sophisticated models of life's earliest molecular systems.
Diagram 2: Emergence of biological systems from unified model (13 words)
From Primordial Soup to Modern Labs: RNA Engineering and Therapeutic Applications
Directed Evolution of RNA Polymerase Ribozymes for Enhanced Replication
The RNA world hypothesis posits that RNA once served as both the primary genetic material and the catalytic molecule in early life, prior to the evolutionary emergence of DNA and proteins [3] [38]. A critical requirement for this hypothesized stage in life's origins is a self-replicating RNA system, capable of Darwinian evolution. The central pillar of this system would be an RNA polymerase ribozyme – an RNA molecule that can catalyze the template-directed copying of RNA sequences, including its own [39] [40]. Such a ribozyme would bridge the gap between inanimate chemistry and an RNA-based biology.
However, naturally occurring self-replicating ribozymes are not known in extant biology, necessitating their de novo creation in the laboratory. Directed evolution has proven to be a powerful methodology for this endeavor, allowing researchers to mimic natural selection in a test tube. This process involves iterative rounds of selection and amplification to isolate ribozyme variants with enhanced catalytic capabilities from large, diverse RNA libraries [39]. This technical guide details the experimental strategies and breakthroughs in the directed evolution of RNA polymerase ribozymes, framing them within the broader context of prebiotic chemistry research and the ongoing quest to validate the RNA world hypothesis. Recent advances demonstrate that evolved ribozymes are now capable of synthesizing complex functional RNAs, including full-length copies of their own evolutionary ancestors, bringing us closer than ever to a functional RNA-based replication system [39].
Key Experimental Breakthroughs in Ribozyme Evolution
The journey toward an efficient RNA polymerase ribozyme began with the isolation of an RNA ligase ribozyme from a vast pool of random RNA sequences. Through successive generations of directed evolution, this ligase was progressively engineered and optimized to function as an RNA-dependent RNA polymerase.
Table 1: Key Evolved RNA Polymerase Ribozymes and Their Capabilities
| Ribozyme Name | Generations of Evolution | Key Features and Innovations | Synthetic Capabilities |
|---|---|---|---|
| Class I Ligase Ancestor | Starting point | Original catalytic RNA | RNA ligation |
| Wild-type Polymerase | Early evolution | Appended accessory domain and processivity tag [39] | Primer extension up to 14 NTPs [39] |
| Z RPR | Further optimization | -- | Primer extension up to 20 NTPs [41] |
| 24-3 Polymerase | 24 rounds from wild-type | Improved activity on structured templates [39] | Synthesis of hammerhead ribozyme [39] |
| 38-6 Polymerase | 38 rounds from wild-type | ~10-fold higher activity than 24-3 [39] | Synthesis of yeast phenylalanyl-tRNA [39] |
| 52-2 Polymerase | 52 rounds from wild-type | Structural rearrangement of catalytic core; novel pseudoknot [39] | 23-fold more efficient synthesis of class I ligase [39] |
| Triplet Polymerase (TPR) | Evolved from Zcore RPR | Uses trinucleotide triphosphates (triplets); heterodimeric structure [40] | Copies highly structured templates, including its own catalytic domain [40] [42] |
A landmark achievement in this field was the structural evolution of the polymerase core. Starting from the 38-6 polymerase, 14 additional rounds of evolution under increasingly stringent conditions (including reduced Mg²⁺ concentration to potentially enhance fidelity) yielded the 52-2 polymerase [39]. Deep sequencing of the evolutionary trajectory revealed that the population explored multiple paths, converging on a solution involving a tertiary structural rearrangement. This innovation involved 11 substitutions, 2 insertions, and 2 deletions, which shortened an existing stem and formed a new one, creating a novel pseudoknot structure near the active site [39]. This demonstrates that directed evolution can drive significant structural innovation, not just local refinement, allowing the ribozyme to escape local fitness peaks.
A parallel breakthrough addressed the fundamental "structure vs. replication" paradox: the fact that functional RNAs require stable folded structures, yet these same structures block their own replication by polymerase ribozymes [40]. A radical solution emerged from rethinking the building blocks of replication. Researchers evolved a triplet polymerase ribozyme (TPR) that uses 5'-triphosphorylated RNA trinucleotides (triplets) instead of single nucleotides (NTPs) [40]. This heterodimeric ribozyme, emerging from in vitro evolution as a mutualistic RNA heterodimer, exhibits several transformative properties:
- Cooperative strand invasion: Triplets bind tightly and cooperatively to the template, invading and unraveling stable secondary structures that are intractable to canonical polymerase ribozymes [40].
- Bidirectional and primer-free synthesis: The TPR supports non-canonical modes of synthesis, including initiation without a pre-formed primer [40].
- Rolling circle synthesis (RCS): The TPR can catalyze continuous RNA synthesis on small circular RNA templates, driving strand displacement to produce long, single-stranded concatemeric products. This process offers a potential solution to the strand separation problem in RNA replication [42]. Furthermore, all steps of a viroid-like replication pathway—RCS, product cleavage, and re-circularization—have been demonstrated to be catalyzed by RNA alone [42].
Detailed Experimental Protocols
This section provides detailed methodologies for key experiments in the directed evolution and characterization of RNA polymerase ribozymes.
General Directed Evolution Workflow for Ribozymes
The following protocol outlines the core cycle of directed evolution, as applied to develop polymerases like the 52-2 variant and the TPR [39] [40].
1. Library Generation:
- Starting Point: Begin with a parent ribozyme sequence (e.g., the 38-6 polymerase).
- Diversification: Introduce random mutations throughout the sequence using error-prone PCR. A typical mutation rate is 10% per nucleotide position [39]. For the TPR evolution, a library of 1.5 × 10¹⁵ Zcore variants with a new random 3' N30 region was used [40].
2. In Vitro Transcription:
- Transcribe the DNA library in vitro to generate the corresponding pool of RNA ribozyme variants.
3. Selection:
- Incubate the RNA pool under conditions that favor the desired activity. For polymerase evolution, this typically involves:
- Template: Providing an RNA template to be copied (e.g., a hammerhead ribozyme sequence).
- Substrates: Providing activated substrates (NTPs or triplet triphosphates).
- Conditions: Often employing the eutectic phase of water ice to enhance RNA stability and ribozyme activity by reducing water activity and concentrating reactants [40] [42].
- The selection criterion is engineered so that ribozymes which successfully perform the templated synthesis become physically tagged (e.g., covalently linked to a primer) or otherwise separable from inactive variants [40].
4. Recovery and Amplification:
- Isolate the RNA from successful ribozymes.
- Reverse Transcribe the RNA into cDNA.
- Amplify the cDNA using PCR (or error-prone PCR to maintain diversity for the next round).
5. Iteration:
- The resulting DNA library is used to start the next round of evolution. This cycle is repeated for dozens of rounds, with selection pressure gradually intensified (e.g., by reducing reaction time, substrate concentration, or Mg²⁺ levels) to drive improvement [39].
Protocol for Assessing Ribozyme Activity via Quantitative RT-PCR
Accurately measuring polymerization activity is crucial. While traditional methods use gel electrophoresis and densitometry, quantitative RT-PCR provides a sensitive and reliable alternative [43].
Materials:
- Purified ribozyme RNA
- RNA template and primer
- Substrates (NTPs or triplet triphosphates)
- Reaction buffer (often including MgCl₂)
- Reverse transcriptase, primers, and fluorescent dyes for qPCR.
Method:
- Cleavage Reaction: Incubate the ribozyme with its template and substrates for a defined time (e.g., up to 3600 seconds).
- Reaction Termination: Heat-inactivate the ribozyme.
- Quantitative RT-PCR:
- Use the reacted mixture as a template for one-step quantitative RT-PCR.
- Design primers to amplify a region of the original substrate RNA.
- The amount of remaining full-length substrate RNA is inversely proportional to the ribozyme's cleavage/polymerization activity.
- Data Analysis: Calculate the fraction of uncleaved substrate (Su) at a given time (e.g., Su3600). Ribozyme activity can be expressed as (1 - Su3600). This value has been shown to be highly consistent with results from gel electrophoresis and densitometry [43].
Protocol for Triplet-Based Rolling Circle Synthesis (RCS)
This protocol leverages the TPR to replicate circular RNA templates, addressing the strand separation problem [42].
Materials:
- Triplet Polymerase Ribozyme (TPR)
- Small circular RNA (scRNA) template
- RNA primer (complementary to a region on the circle)
- Trinucleotide triphosphates (pppNNN)
- Eutectic phase reaction buffer.
Method:
- Hybridization: Anneal the RNA primer to the scRNA template.
- RCS Reaction:
- Combine the primer-template complex with the TPR and triplet triphosphates.
- Incubate in the eutectic phase of water ice to facilitate the reaction.
- Strand Displacement: The TPR catalyzes the iterative ligation of triplets. The binding of subsequent triplets cooperatively invades the duplex, displacing the nascent strand's 5'-end. This results in continuous, primer-driven synthesis around the circle, producing long, single-stranded concatemeric RNA products.
- Analysis: Analyze products by denaturing gel electrophoresis to visualize concatemers of various lengths, confirming successful RCS and strand displacement.
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Research Reagents for Ribozyme Evolution and Characterization
| Reagent / Tool | Function and Rationale | Example Use Case |
|---|---|---|
| Error-Prone PCR | Introduces random mutations into ribozyme gene libraries to create genetic diversity for selection. | Generating the initial diverse pool of ribozyme variants from a parent sequence [39]. |
| Eutectic Ice Phases | A reaction medium where water ice coexists with concentrated liquid brine. Enhances RNA stability and ribozyme activity by reducing hydrolysis and concentrating reactants [40]. | Critical reaction environment for the activity of the Triplet Polymerase Ribozyme (TPR) [40] [42]. |
| Trinucleotide Triphosphates (pppNNN) | Activated RNA trimers used as polymerization substrates. Enable cooperative invasion of structured RNA templates and facilitate strand displacement. | Substrate for TPR in copying structured RNAs and performing Rolling Circle Synthesis [40] [42]. |
| Small Circular RNA (scRNA) Templates | Topologically closed RNA templates that enable Rolling Circle Synthesis. Avoid free ends that can lead to unproductive duplex formation. | Template for demonstrating continuous, strand-displacing RNA synthesis by the TPR [42]. |
| In Vitro Transcription Kits | Generate large quantities of RNA ribozymes and templates from DNA for use in selection rounds and biochemical assays. | Production of ribozyme libraries and target templates (e.g., hammerhead ribozyme) for selection experiments. |
| Deep Sequencing | High-throughput sequencing of entire populations of evolved ribozymes. Allows for tracking evolutionary trajectories and identifying beneficial mutations. | Mapping the evolutionary path from the 38-6 to the 52-2 polymerase, revealing structural remodeling [39]. |
Visualization of Ribozyme Mechanisms and Workflows
Triplet-Mediated Strand Invasion in RNA Replication
The unique properties of triplet substrates resolve the structure-replication paradox by enabling the ribozyme to copy through stable secondary structures.
A Viroid-Like RNA Replication Cycle Catalyzed by RNA
This diagram outlines a potential pathway for autonomous RNA replication, where all steps are catalyzed by RNA, as demonstrated in recent experiments [42].
Directed evolution has successfully transformed simple RNA ligases into increasingly sophisticated RNA polymerase ribozymes. The field has moved from synthesizing short oligonucleotides to producing complex, functional RNAs like the hammerhead ribozyme and tRNA. Two key innovations—tertiary structural remodeling of the catalytic core and the adoption of triplet-based replication—have overcome major hurdles, demonstrating that evolution can discover non-intuitive solutions to fundamental problems like template structure and strand separation.
These advances provide experimental plausibility for key aspects of the RNA world hypothesis, showing that RNA can, in principle, catalyze its own replication. The development of a viroid-like replication cycle, entirely catalyzed by RNA, marks a significant step toward a minimal self-sustaining system [42]. Future research will focus on integrating these breakthroughs into a single, robust ribozyme capable of accurate, processive self-replication under prebiotically plausible conditions. This work not only illuminates potential pathways for the origin of life but also pushes the boundaries of synthetic biology, with potential applications in biotechnology and therapeutics.
The emergence of artificial intelligence (AI) is revolutionizing our ability to model RNA structure and function, bridging fundamental research and therapeutic development. This whitepaper provides an in-depth technical analysis of cutting-edge AI methodologies that predict RNA three-dimensional folding and interactions with small molecule ligands. We frame these computational advances within the context of the RNA World hypothesis, exploring how modern AI tools provide a new lens to study prebiotic chemistry and the molecular origins of life. For researchers and drug development professionals, this guide details experimental protocols, compares model performance, and presents essential research tools, underscoring how AI-driven insights into RNA's dual role as an information carrier and catalyst are expanding the druggable genome.
The RNA World hypothesis posits that early life was based on RNA molecules, which served both as the repository of genetic information and the catalytic engine for biochemical reactions [23] [44]. This hypothesis is grounded in RNA's unique capacity for information storage, self-replication, and catalysis—properties that modern AI modeling is now revealing with unprecedented atomic detail. While only about 1.5% of the human genome codes for proteins, approximately 70% is transcribed into non-coding RNA (ncRNA), presenting a vast landscape of potential therapeutic targets that remain largely unexplored [45].
Traditional drug discovery has focused predominantly on protein targets, with an estimated 90% of marketed drugs being small molecules that interact with proteins [46]. However, many disease-relevant proteins are considered "undruggable" due to the absence of suitable binding pockets. RNA structures offer an alternative targeting strategy, but their highly dynamic and complex nature has made them resistant to conventional structural determination and modeling approaches [47].
The integration of artificial intelligence into RNA structural biology is overcoming these historical limitations. AI models are now capable of predicting RNA tertiary structures from sequence data, identifying functional binding sites, and scoring small molecule interactions with therapeutic potential—all while accounting for the dynamic conformational ensembles that characterize functional RNA molecules [48] [49].
AI Methodologies for RNA Structure Prediction
From Sequence to Structure: Deep Learning Architectures
Machine learning, particularly deep learning (DL), has emerged as the predominant approach for tackling the challenges of RNA structure prediction. These methods have evolved from early thermodynamic models that predicted minimum free-energy structures to sophisticated neural networks that learn complex sequence-structure relationships from growing repositories of experimental data [47].
The core challenge lies in RNA's structural hierarchy:
- Primary structure: The linear nucleotide sequence (A, U, G, C)
- Secondary structure: Canonical base pairs (A-U, G-C, G-U) forming helices and motifs
- Tertiary structure: The three-dimensional arrangement of these elements in space [49]
Different AI architectures excel at different levels of this hierarchy. Graph Neural Networks (GNNs) process molecular structures as mathematical graphs where atoms serve as nodes and bonds as edges, making them particularly suited for modeling tertiary interactions and molecular surfaces [50] [46]. Convolutional Neural Networks (CNNs), originally developed for image processing, have been adapted to recognize spatial patterns in RNA structural data, while transformer-based models capture long-range interactions within nucleotide sequences that influence folding patterns [47].
Table 1: AI Model Architectures for RNA Structure Prediction
| Model Type | Primary Application | Key Features | Limitations |
|---|---|---|---|
| Graph Neural Networks (GNNs) | Tertiary structure, binding site prediction | Processes molecular graphs; captures atomic-level interactions | Requires substantial computational resources |
| Convolutional Neural Networks (CNNs) | Secondary structure motifs | Recognizes spatial patterns in sequence data | Limited capacity for long-range interactions |
| Transformer-based Models | Sequence-structure relationships | Captures dependencies across entire sequence | High data requirements for training |
| Geometric Deep Learning | Molecular surface interactions | Models 3D shape and chemical features | Dependent on quality of structural templates |
Integrating Experimental Data with AI Prediction
Even the most advanced AI models face challenges in achieving high accuracy due to the limited availability of high-resolution RNA structures. Integrative approaches that combine computational predictions with experimental data are emerging as powerful solutions to this problem.
The SCOPER (SOlution Conformation PrEdictor for RNA) pipeline, developed at Lawrence Berkeley National Laboratory, exemplifies this trend by combining AI-based structure prediction with Small Angle X-Ray Scattering (SAXS) experimental data [48]. This methodology begins with initial structure predictions from tools like AlphaFold3, then refines these models using SAXS data that provides information about RNA conformation in solution. A key innovation in SCOPER is the machine learning component that accurately places magnesium ions—critical for RNA folding stability—within the structural model [48].
This hybrid approach addresses a fundamental limitation of purely computational methods: the dynamic nature of RNA molecules that often adopt multiple conformations in equilibrium. As Michal Hammel, a staff scientist at Berkeley Lab, notes: "These days, programs like AlphaFold are almost 95% accurate for proteins but much worse for RNA. It will sometimes come up with five different models that are different. And now the question is, which one is right? SCOPER can tell you" [48].
Predicting RNA-Small Molecule Interactions
Data-Driven Scoring of Molecular Interactions
The prediction of how small molecules interact with RNA targets represents one of the most promising applications of AI in drug discovery. The RNAsmol framework, developed by Zhi John Lu's team at Tsinghua University, demonstrates how innovative training strategies can overcome the limited availability of RNA-small molecule interaction data [45].
RNAsmol employs a deep learning approach that combines data perturbation and data augmentation strategies. Data perturbation introduces controlled variations to training data, simulating the diversity encountered in real screening environments and improving model robustness. Data augmentation generates virtual negative samples and potential unlabeled samples based on known interactions, expanding the model's ability to identify novel binding compounds [45].
This approach has demonstrated significant performance improvements, with the average AUROC (Area Under the Receiver Operating Characteristic) increasing by approximately 8% in 10-fold cross-validation, and performance on unseen samples improving by about 16% compared to traditional methods [45]. In virtual screening applications, RNAsmol improved ranking scores by approximately 30% when distinguishing between bait molecules and real ligands, highlighting its potential for identifying genuine binding partners amid diverse chemical libraries [45].
Geometric Deep Learning for Binding Site Prediction
Accurate prediction of binding sites is prerequisite to designing effective small molecule therapeutics. The RLBSIF (RNA-Ligand Binding Surface Interaction Fingerprints) method introduces a geometric deep learning approach that characterizes RNA-ligand interactions through molecular surface features [50].
This method utilizes surface geometric features (shape index and distance-dependent curvature) combined with chemical features (atomic charge) to create comprehensive interaction fingerprints. The ResNet18 network then analyzes these fingerprints to identify ligand binding pockets. Trained on 440 binding pockets, RLBSIF achieves a remarkable overall pocket-level classification accuracy of 90% and can predict binding sites at nucleotide resolution through a full-space enumeration method [50].
Table 2: Performance Comparison of RNA-Small Molecule Interaction Models
| Model | Approach | Key Metrics | Unique Advantages |
|---|---|---|---|
| RNAsmol | Data perturbation & augmentation | 8% improvement in AUROC; 30% better ligand ranking | Predicts from sequence alone; no 3D structure required |
| RLBSIF | Geometric deep learning & surface fingerprints | 90% binding site classification accuracy | Nucleotide-resolution binding site prediction |
| Boltz-2 | Binding affinity prediction | 20-second calculation per prediction | Thousand times faster than physical simulations |
| Hermes | Sequence/SMILES-based binding prediction | 200-500x faster than Boltz-2 | Trained on proprietary high-quality dataset |
Experimental Protocols and Methodologies
Protocol: Virtual Screening with RNAsmol
The following protocol outlines the standard workflow for virtual screening of small molecule libraries against RNA targets using the RNAsmol framework [45]:
Data Preparation:
- Obtain the RNA target sequence in FASTA format
- Prepare small molecule structures in SMILES (Simplified Molecular Input Line Entry System) format
- For known active compounds, create a curated set of positive examples
- Generate decoy molecules using data perturbation strategies
Feature Representation:
- Encode RNA sequences using RNA-specific grammatical rules (A-U, G-C, G-U pairs)
- Represent small molecules as graph structures with atomic nodes and bond edges
- Extract molecular features using graph diffusion convolution modules
Model Training:
- Implement data augmentation by generating virtual negative samples
- Apply perturbation to training data to simulate real-world diversity
- Train the deep learning model with multi-modal feature fusion
- Use attention mechanisms to weight integrated RNA and small molecule features
Interaction Scoring:
- Input query RNA-small molecule pairs into the trained model
- Generate interaction scores predicting binding likelihood
- Rank compounds based on predicted affinity
Validation:
- Perform 10-fold cross-validation to assess model performance
- Evaluate on hold-out test sets of unseen RNA-small molecule pairs
- Compare against known active and inactive compounds using AUROC metrics
Protocol: Binding Site Identification with RLBSIF
This protocol details the process for identifying RNA-small molecule binding sites using the RLBSIF geometric deep learning approach [50]:
Structure Preparation:
- Obtain RNA 3D structure from PDB or computational prediction
- Preprocess structure to add missing atoms and optimize hydrogen bonds
- Ensure structural integrity and correct non-canonical base pairs
Surface Characterization:
- Generate molecular surface using marching cubes algorithm
- Calculate shape index and distance-dependent curvature at each surface point
- Compute atomic partial charges using empirical methods
Fingerprint Generation:
- Create interaction fingerprints by combining geometric and chemical features
- Align fingerprints to reference binding sites for comparison
- Apply dimensionality reduction to highlight discriminative features
Binding Site Prediction:
- Input surface fingerprints into pre-trained ResNet18 network
- Classify potential binding pockets using full-space enumeration
- Generate nucleotide-resolution predictions of ligand interaction sites
Experimental Validation:
- Compare predictions with known binding sites from structural databases
- Validate predictions through mutagenesis studies or competitive binding assays
- Refine model based on experimental feedback
Virtual Screening Workflow for RNA-Targeted Small Molecules
Table 3: Essential Research Reagents and Computational Tools for AI-Driven RNA Research
| Resource | Type | Function | Access |
|---|---|---|---|
| SCOPER | Computational Pipeline | Integrates SAXS data with AI predictions for RNA structure determination | Open-source [48] |
| RNAsmol | Deep Learning Model | Scores RNA-small molecule interactions from sequence data | Available upon publication [45] |
| RLBSIF | Geometric Deep Learning | Predicts RNA-small molecule binding sites from 3D structure | GitHub: ZUSTSTTLAB/RLBSIF [50] |
| SAIR Repository | Structural Database | Computationally folded protein-ligand structures with affinity data | Open-access [51] |
| Boltz-2 | Binding Affinity Model | Predicts small molecule binding affinity in seconds | MIT License [51] |
| ChEMBL/BindingDB | Experimental Database | Curated binding affinity data for model training | Public [51] |
| SIBYLS Beamline | SAXS Instrumentation | Provides solution-state RNA structural data | ALS User Facility [48] |
Implications for the RNA World Hypothesis
AI-driven RNA modeling is providing unprecedented insights into the RNA World hypothesis by demonstrating how simple molecular languages could have driven the emergence of biological complexity. As Professor Zhi John Lu's team notes: "We tried to use a simple, RNA-specific grammar (such as A-U, G-C, G-U) to represent RNA molecules... This specific and simple grammar not only achieved unexpected results in the above work, but also made us more convinced of an academic hypothesis that is familiar in the RNA field: the origin of life is a RNA world" [45].
Recent experimental work supporting the RNA World hypothesis has discovered RNA enzymes capable of accurately replicating RNA strands while allowing for the emergence of new molecular variants over time [44]. Senior author Gerald Joyce of the Salk Institute reflects that "we're chasing the dawn of evolution. By revealing these novel capabilities of RNA, we're uncovering the potential origins of life itself, and how simple molecules could have paved the way for the complexity and diversity of life we see today" [44].
The study of non-canonical nucleotides—approximately 170 variants beyond the standard A, U, G, and C—provides additional clues about prebiotic evolution. These modified nucleotides may have played crucial roles in early RNA world scenarios, facilitating the emergence of catalytic RNAs and the origin of template-directed synthesis [52].
RNA World Hypothesis and Molecular Evolution
The field of AI-driven RNA modeling is progressing rapidly, with several emerging trends shaping its future trajectory. Agentic AI systems that can autonomously navigate discovery pipelines are showing promise for identifying drug candidates, particularly in oncology and immunology [51]. The integration of physics-based simulations with data-driven models addresses the critical need for incorporating biophysical principles into AI predictions [49] [51].
For the RNA World hypothesis, these computational advances offer new opportunities to test long-standing questions about life's origins. As researchers create increasingly sophisticated RNA models that replicate, evolve, and catalyze reactions, we move closer to understanding how life could emerge from simple molecular systems. The "seemingly simple RNA language," as described by Lu's team, may indeed contain "the basic elements of the origin of life or even the origin of the universe: information replication, transmission and mutation" [45].
In conclusion, AI-driven RNA modeling represents a transformative approach to understanding both fundamental biology and therapeutic development. By revealing the structural principles and interactive capabilities of RNA, these methods provide powerful tools for drug discovery while simultaneously illuminating the ancient molecular processes that may have given rise to life itself.
The pursuit of synthetic biological systems that operate predictably within living cells is a central goal of synthetic biology. A significant challenge in this field is that engineered genetic circuits often face inadvertent interference from the host's native machinery, which can compromise their function and reduce host fitness [53]. To overcome this, researchers are developing orthogonal systems—biological components and circuits that are insulated from host processes, thereby functioning independently and reliably [53]. This concept of biological orthogonalization finds a profound historical parallel in the RNA World Hypothesis, which proposes that early life was based on RNA molecules capable of both storing genetic information and catalyzing chemical reactions, a self-contained system operating before the advent of DNA and proteins [23].
Modern synthetic biology leverages these principles to construct RNA-based regulatory systems. Unlike protein-based circuits, RNA circuits offer advantages such as faster design cycles, reduced metabolic burden on the host, and simpler thermodynamic predictability [54] [55]. By designing synthetic RNA systems that minimize cross-talk with host machinery, researchers are essentially creating modern analogues of the primordial RNA world, enabling precise control over cellular behavior for applications in therapeutics, biosensing, and bio-production [53] [56]. This technical guide explores the core principles, components, and methodologies for building such orthogonal biological circuits using synthetic RNA systems.
Core Principles of RNA-Based Orthogonal Systems
The Concept of Biological Orthogonalization
In synthetic biology, orthogonality describes the inability of two or more biomolecules, similar in composition or function, to interact with one another or affect one another's substrates [53]. For example, two orthogonal proteases would be unable to cleave each other's target sequences. The primary objective is to create a user-controlled paralogue of the central dogma—a system for information storage, transfer, and translation that operates alongside, but independently of, the host's native processes [53]. This is crucial because engineered circuits that heavily repurpose host machinery can deplete essential resources, cause toxicity, and exhibit unpredictable performance due to unintended interactions [53]. Orthogonalization mitigates these issues, enhancing circuit reliability and context-independent functionality.
Advantages of RNA as an Orthogonal Substrate
RNA molecules serve as ideal substrates for building orthogonal circuits due to several key properties:
- Programmability and Predictability: RNA-RNA interactions are governed by Watson-Crick base pairing, making their thermodynamics highly predictable and their design straightforward [54] [55].
- Fast Kinetics and Reduced Burden: RNA-based regulators exhibit faster signal propagation and degradation rates compared to proteins. They also place less burden on cellular resources as they do not require translation [54] [55].
- Direct Information Processing: RNA circuits can directly propagate regulatory signals as RNA molecules, eliminating the need for protein intermediaries and simplifying circuit architecture [55].
- Safety in Therapeutics: For medical applications, all-RNA gene circuits are transient and do not cause insertional mutagenesis, making them a safer option for cell and gene therapies [56].
Key RNA Components for Circuit Engineering
Synthetic biologists have developed a versatile toolbox of de novo-designed RNA regulators. The table below summarizes the core components used to construct orthogonal RNA circuits.
Table 1: Key Synthetic RNA Regulators for Orthogonal Circuits
| Component Name | Type | Mechanism of Action | Key Features |
|---|---|---|---|
| Small Transcriptional Activating RNA (STAR) [54] | Transcriptional Regulator | Binds to target RNA to prevent terminator formation, allowing transcription. | Provides activation at the transcriptional level; enables complex logic gates. |
| Toehold Switch (THS) [54] | Riboregulator | Sequesters RBS and start codon; trigger RNA binding unwinds the structure, initiating translation. | High ON/OFF ratios; modular; enables multi-input logic processing. |
| Three-Way Junction (3WJ) Repressor [54] | Translational Repressor | Forms a stable three-way junction upon trigger binding, inhibiting translation. | Functions as a repressor; can be combined with toehold switches for NAND/NOR logic. |
| CaVT (Caliciviral VPg-based Translational Activator) [56] | Translational Activator | Uses viral VPg protein and RNA-binding domains to directly activate translation of target mRNAs. | First synthetic tool for direct translational activation in human cells; reduces circuit complexity. |
| pT181-Derived Attenuator [55] | Transcriptional Regulator | Antisense RNA-mediated transcription attenuation mechanism that controls plasmid copy number. | Enables orthogonal variant creation; captures key regulatory features in a single molecule. |
The following diagram illustrates the operational mechanisms of three fundamental RNA regulators: the Toehold Switch, the STAR system, and the 3WJ Repressor.
Implementing Dynamic Control: RNA-Based Circuit Motifs
The Incoherent Feed-Forward Loop (IFFL)
A powerful network motif for generating dynamic responses is the Type 1 Incoherent Feed-Forward Loop (IFFL). This motif produces pulsed output in response to a persistent input and has applications in biosensing, fold-change detection, and maintaining constant expression levels [54]. In an IFFL, an input (X) directly activates an output (Z) while also activating a repressor (Y) that inhibits Z. The delay in the repression arm, due to the time required to produce Y, results in a pulse of Z expression [54].
Researchers have implemented IFFLs using different combinations of RNA and protein components:
- RNA-Only IFFL: Utilizes a STAR system for direct activation and a 3WJ repressor for inhibition. While fast, its rapid RNA-RNA interaction kinetics often lack the significant timescale difference needed for a pronounced pulse [54].
- RNA-Protein Hybrid IFFL: Employs a Toehold Switch for activation and a protein (e.g., TetR) for inhibition. The slower synthesis and maturation of the protein introduce the necessary delay for robust pulse generation [54].
The diagram below contrasts the architectures and behaviors of these two IFFL implementations.
Quantitative Performance of IFFL Circuits
The dynamic behavior of IFFL circuits is characterized by key kinetic parameters. The data below, derived from mechanistic modeling and experimental validation, highlights the performance differences between RNA-only and RNA-protein hybrid IFFLs [54].
Table 2: Kinetic Parameters for IFFL Circuit Designs
| Parameter | RNA-Only IFFL (STAR & 3WJ) | RNA-Protein Hybrid IFFL (THS & TetR) |
|---|---|---|
| Activation Pathway Delay | Minimal (Fast RNA-RNA binding) | Minimal (Fast RNA-RNA binding) |
| Repression Pathway Delay | Minimal (Fast RNA-RNA binding) | Significant (Slow protein synthesis & maturation) |
| Timescale Difference | Insufficient for pronounced pulsing | Sufficient for robust pulsing |
| Pulse Generation | Not observed experimentally | Observed over a wide inducer concentration range |
| Model-Guided Design | ODE models confirmed lack of pulse | ODE models predicted successful pulse generation |
Experimental Protocol: Implementing an RNA-Protein Hybrid IFFL
This section provides a detailed methodology for constructing and testing the RNA-protein hybrid IFFL circuit in E. coli, as validated by research [54].
Plasmid Construction and Bacterial Strains
- Plasmid Backbones: Use compatible commercial vectors with different antibiotic resistance and origins of replication (e.g., pET15b, pCDFDuet, pCOLADuet) to harbor different circuit nodes and avoid plasmid incompatibility [54].
- Circuit Node Cloning:
- Node X (Input): Clone the gene for the toehold trigger (input sensor) into the pET15b vector.
- Node Y (Repressor): Clone the gene for the TetR repressor protein under the control of a toehold switch into the pCDFDuet vector.
- Node Z (Output): Clone the gene for the output reporter (e.g., GFPmut3b) under the control of a TetR-repressible promoter (e.g., P_{LtetO-1}) into the pCOLADuet vector.
- Assembly: Perform cloning via Gibson assembly or similar methods. Verify all constructs by DNA sequencing after plasmid purification [54].
- Host Strain: Transform the finalized plasmids into an appropriate E. coli strain such as BL21 DE3 for the RNA-protein hybrid circuit [54].
Cell Culture and Induction
- Starter Culture: Inoculate a single transformed colony into 500 µL of LB liquid medium supplemented with the appropriate antibiotics (Ampicillin, Spectinomycin, Kanamycin). Grow overnight (~16 hours) in a 96-well plate with shaking at 800 rpm and 37°C [54].
- Experimental Culture: Dilute the overnight culture 1/100-fold into fresh, antibiotic-supplemented LB medium.
- Induction: Immediately treat the diluted cultures with the chemical inducer (e.g., IPTG) across a range of concentrations (e.g., 500 µM, 125 µM) to activate the circuit [54].
Data Collection and Analysis
- Time-Course Measurement: Use a microplate reader to measure the fluorescence (output Z) and optical density (cell growth) over time.
- Pulse Identification: Analyze the fluorescence data to identify the characteristic pulse dynamics—a rapid increase in output followed by a decline as the repressor protein accumulates.
- Model Validation: Fit the experimental data to ordinary differential equation (ODE)-based mathematical models to validate circuit performance and refine kinetic parameters [54].
The Scientist's Toolkit: Essential Research Reagents
The table below lists key reagents and tools required for the design and implementation of synthetic RNA circuits.
Table 3: Research Reagent Solutions for Synthetic RNA Circuits
| Reagent/Tool | Function/Description | Example Use Case |
|---|---|---|
| Orthogonal Polymerases/Replication Systems [53] | Replicates orthogonal genetic information independently of the host genome. | Cytoplasmic plasmid systems (e.g., OrthoRep in yeast) for stable circuit maintenance. |
| Non-Canonical Nucleotides [53] [52] | Modified nucleobases (e.g., m6dA, phosphorothioates) that insulate synthetic DNA/RNA from host nucleases and machinery. | Creating epigenetic orthogonal control systems [53] or studying prebiotic evolution [52]. |
| Synthetic RNA Spike-in Controls (ERCC RNAs) [57] | Exogenous RNA sequences with known concentrations used to calibrate and assess the sensitivity and accuracy of RNA-seq experiments. | Quantifying transcript abundance and detecting biases in RNA-seq data from circuits [57]. |
| Mechanistic Modeling (ODE Models) [54] | Mathematical models based on ordinary differential equations that simulate circuit dynamics. | Predicting circuit performance, debugging failures, and guiding optimal design before experimental implementation [54]. |
| Viral Translational Activators (e.g., VPg) [56] | Viral proteins adapted to directly initiate translation on synthetic mRNAs in mammalian cells. | Simplifying circuit design in human cells (e.g., CaVT system) for therapeutic applications [56]. |
Connecting to the RNA World and Prebiotic Chemistry
The engineering of orthogonal RNA circuits resonates deeply with the RNA World Hypothesis. This hypothesis posits that early life relied on RNA for both genetic information storage and catalytic function, a primordial form of biological orthogonality that existed before the complex interplay of DNA, RNA, and proteins evolved [23]. Modern synthetic RNA circuits can be viewed as a reductionist attempt to recreate simplified, engineerable versions of this ancient world within modern cells.
Research into non-canonical nucleotides further bridges synthetic biology and prebiotic chemistry. Natural systems contain about 170 different modified RNA nucleotides [52]. These modifications, such as N6-methyldeoxyadenosine (m6dA) and 5-Methylcytosine, are now understood not just as epigenetic regulators but also as potential relics of a more diverse prebiotic chemical landscape [53] [52]. Their study informs the design of synthetic nucleotides that can expand the genetic alphabet and form the basis of highly orthogonal systems, much as they might have in the origins of life [53] [52]. The drive to create insulated circuits using synthetic polymers mirrors the evolutionary transition from a world of competing molecular systems to the consolidated biological central dogma we know today.
The RNA world hypothesis posits that early life on Earth was based on RNA molecules capable of both storing genetic information and catalyzing chemical reactions, predating the DNA-protein world we observe today [23]. This primordial role of RNA as a multifunctional molecule finds a modern parallel in the rapidly advancing field of RNA-targeting therapeutics. By targeting RNA, researchers are developing precise interventions for previously "undruggable" diseases, effectively harnessing RNA's central role in gene expression [58] [59].
Current RNA therapeutics leverage diverse mechanisms including direct gene silencing, splice modulation, and therapeutic protein expression. This review provides a comprehensive technical analysis of two major therapeutic classes: small interfering RNA (siRNA) and small molecule splicing modulators, framing them within the context of prebiotic chemistry while providing practical experimental guidance for research and development professionals.
siRNA Therapeutics: Mechanism and Clinical Advancement
Molecular Mechanisms and Delivery Challenges
Small interfering RNA (siRNA) therapeutics harness the natural RNA interference (RNAi) pathway to achieve highly specific gene silencing. The mechanism begins with synthetic double-stranded siRNA incorporation into the RNA-induced silencing complex (RISC). The complex's catalytic component, Argonaute-2 (Ago-2), cleaves and releases the passenger strand, allowing the guide strand to direct RISC to complementary mRNA sequences through Watson-Crick base pairing. Upon binding, Ago-2 mediates target mRNA cleavage, preventing translation and enabling degradation of the mRNA fragments [59].
Despite this elegant mechanism, siRNA therapeutics face substantial delivery barriers that have slowed clinical translation. Key challenges include rapid renal clearance (with naked siRNA having a half-life as short as 5 minutes), degradation by serum and tissue nucleases, inefficient cellular uptake due to large size and negative charge, and endosomal trapping where less than 1% of internalized siRNA reaches the cytoplasmic target site [59] [60]. Additionally, unintended immune recognition through Toll-like receptors can trigger inflammatory responses [59].
Chemical Modifications and Delivery Strategies
To overcome these barriers, extensive chemical modifications have been developed:
- Phosphorothioate (PS) backbone: Replacing non-bridging oxygen with sulfur increases nuclease resistance and promotes plasma protein binding, extending circulation half-life [59].
- 2'-sugar modifications: 2'-fluoro (2'-F), 2'-O-methyl (2'-OMe), and 2'-O-methoxyethyl (2'-MOE) substitutions protect against endoribonuclease degradation [59].
- GalNAc conjugation: N-acetylgalactosamine enables targeted delivery to hepatocytes via asialoglycoprotein receptor-mediated endocytosis [59].
Advanced delivery systems are crucial for tissue-specific targeting:
- Lipid nanoparticles (LNPs): Ionizable lipids form nanoparticles that encapsulate siRNA, enhance stability, and facilitate endosomal escape through the proton sponge effect [60].
- Mesoporous silica nanoparticles (MSNs): Offer high loading capacity, protection against degradation, and surface functionalization for targeted delivery [60].
- Polymeric carriers: Biodegradable polymers provide controlled release and reduced immunogenicity [60].
Table 1: Clinically Approved siRNA Therapeutics (2018-2024)
| Drug Name | Target | Indication | Approval Year | Key Technology |
|---|---|---|---|---|
| Patisiran | Transthyretin (TTR) | Hereditary transthyretin-mediated amyloidosis | 2018 | LNP delivery system |
| Givosiran | Aminolevulinic acid synthase 1 (ALAS1) | Acute hepatic porphyria | 2019 | Enhanced stabilization chemistry (ESC)-GalNAc conjugate |
| Lumasiran | Hydroxyacid oxidase 1 (HAO1) | Primary hyperoxaluria type 1 | 2020 | GalNAc conjugate |
| Inclisiran | Proprotein convertase subtilisin/kexin type 9 (PCSK9) | Hypercholesterolemia | 2021 | GalNAc conjugate |
Clinical Landscape and Trial Design Considerations
The clinical development of siRNA therapeutics has accelerated, with 424 clinical trials conducted globally between 2004-2024. Analysis reveals that non-oncology applications dominate (90% of trials), peaking in 2021 with 64 trials, and yielding 6 approved drugs for metabolic and genetic diseases [61]. Key non-oncology targets include PCSK9 for cholesterol management and hepatitis B virus (HBV) genes [61].
In contrast, oncology applications face greater hurdles. Oncology trials initiated later and remain primarily in early phases (60% Phase I), focusing on solid tumors (40%) with target homogenization (40% targeting CSF2) and experiencing a high termination rate (28%) [61]. Cross-target analysis has identified PTGS2 and TGFB1 as shared targets, suggesting potential for combination therapy approaches [61].
Trial design considerations for siRNA therapeutics should account for:
- Dosing frequency: Leveraging siRNA's durable effects (e.g., inclisiran requires biannual dosing) [59]
- Biomarker selection: Monitoring target engagement through mRNA or protein level quantification [61]
- Safety monitoring: Assessing immunogenicity, liver function, and target-related toxicities [59]
Diagram 1: siRNA Mechanism Pathway
Small Molecule Splicing Modulators: From Natural Products to Precision Tools
RNA Splicing Machinery and Cancer-Associated Aberrations
RNA splicing is an essential process in eukaryotic gene expression, involving the precise removal of introns from precursor mRNA (pre-mRNA) and joining of exons to produce mature mRNA. The spliceosome, a massive ribonucleoprotein complex composed of five small nuclear RNAs (U1, U2, U4, U5, U6) and approximately 200 proteins, catalyzes this process through a series of assembly steps (E, A, B, B* complexes) [58].
Alternative splicing allows a single gene to generate multiple mRNA and protein isoforms, with seven major types identified: exon skipping, alternative 5' splice sites, alternative 3' splice sites, intron retention, mutually exclusive exons, alternative promoters, and alternative polyadenylation [58]. In cancer, splicing dysregulation is widespread, with tumors exhibiting up to 30% more alternative splicing events than normal tissues [58]. These aberrations are driven by:
- Mutations in spliceosome components (SF3B1, U2AF1, SRSF2) [58]
- Splice-site-creating mutations that generate novel isoforms [58]
- Dysregulated splicing factor expression (SRSF1, SRSF3, hnRNPA1) [58]
These alterations produce cancer-specific isoforms that drive hallmarks including proliferation, metastasis, angiogenesis, immune evasion, and therapy resistance [58].
Natural Product-Derived Splicing Modulators
The first splicing modulators were discovered through natural product screening:
- Herboxidiene (1992): Isolated from Streptomyces chromofuscus with phytotoxic activity [62]
- FR901464/Spliceostatin family (1996): Identified from Pseudomonas sp. No.2663 as transcriptional activators with antitumor activity [62]
- Pladienolides (2004): Discovered from Streptomyces platensis as angiogenesis inhibitors [62]
Despite structural differences, these compounds share a common molecular target: the SF3B subunit of the U2 snRNP, which is essential for branch point recognition during A complex formation [62]. Structural studies reveal that these compounds adopt similar conformations around their central diene moieties, enabling SF3B1 binding [62].
Table 2: Key Splicing Modulator Classes and Properties
| Compound Class | Origin | Key Derivatives | IC50 (Cell Proliferation) | Molecular Target |
|---|---|---|---|---|
| Pladienolides | Streptomyces platensis | Pladienolide B, E7107 | Low nM range | SF3B1 |
| Spliceostatins | Pseudomonas sp. | FR901464, Spliceostatin A, Meayamycin | Low nM range | SF3B1 |
| Herboxidienes | Streptomyces chromofuscus | Herboxidiene, GEX1A | Low nM range | SF3B1 |
| Synthetic Analogues | N/A | Sudemycins, Branaplam | nM to μM range | SF3B1 |
Therapeutic Applications and Clinical Development
Splicing modulators demonstrate particular promise in oncology, where they can reverse cancer-associated splicing patterns. For example, they can modulate splicing of:
- MCL-1 and BCL-x: Promoting pro-apoptotic isoforms [58]
- VEGF: Altering angiogenic isoforms in ovarian and breast cancers [58]
- CD19: Preventing exon skipping that impairs CAR-T therapy in leukemia [58]
Beyond oncology, splicing modulation shows therapeutic potential for neurological disorders. Branaplam advanced to Phase II trials for Huntington's disease and spinal muscular atrophy, though development was suspended due to nerve damage concerns [63]. Risdiplam (Evrysdi), an FDA-approved orally administered small molecule, treats spinal muscular atrophy by stabilizing the splicing machinery to promote inclusion of exon 7 in the SMN2 gene [63].
Diagram 2: Splicing Modulation Mechanism
Experimental Protocols and Research Tools
Core Methodologies for Splicing Analysis
RNA Sequencing and Alternative Splicing Analysis
- Library Preparation: Use strand-specific RNA-seq protocols with ribosomal RNA depletion to maintain strand orientation and detect non-polyadenylated transcripts [58]
- Sequencing Depth: Minimum 50 million paired-end reads (2x75bp or 2x100bp) per sample to robustly detect alternative splicing events [58]
- Splicing Analysis Pipeline:
- Alignment: Map reads to reference genome using splice-aware aligners (STAR, HISAT2)
- Quantification: Identify splicing events with rMATS, MAJIQ, or LeafCutter
- Visualization: Validate events using IGV or Sashimi plots
- Functional Validation: Confirm biological significance of altered splicing events through:
- Minigene splicing reporters with wild-type and mutant constructs
- qRT-PCR with junction-specific primers to quantify isoform ratios
- Western blotting to detect protein isoform changes [58]
High-Throughput Screening for Splicing Modulators
- Reporter System: Develop cell lines stably expressing dual-luciferase reporters with disease-relevant alternative splicing events (e.g., BCL-x, caspase-9, MNK2) [62]
- Primary Screening: Test compound libraries (10,000-100,000 compounds) at 10μM concentration in 384-well format
- Hit Criteria: >50% splicing modulation at 10μM with <20% cytotoxicity
- Secondary Validation: Dose-response curves (0.1nM-10μM) to determine IC50/EC50 values
- Counter-screens: Exclude non-specific transcription/translation inhibitors [62]
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for RNA-Targeting Therapeutic Research
| Reagent Category | Specific Examples | Function & Application | Key Considerations |
|---|---|---|---|
| siRNA Design Tools | siDirect, BLOCK-iT, Dharmacon siDESIGN | In silico siRNA sequence design and off-target prediction | Guide strand thermodynamics, seed region analysis, genome-wide specificity check |
| Splicing Reporters | pSpliceExpress, pCAS2, minigene constructs | Functional assessment of splicing modulation | Include genomic context with flanking intronic sequences, proper splice site strengths |
| Chemical Modifications | Phosphorothioate, 2'-OMe, 2'-F, 2'-MOE, LNA | Enhance stability, specificity, and pharmacokinetics | Balance modification density with RISC compatibility and toxicity |
| Delivery Systems | Lipofectamine RNAiMAX, TransIT-mRNA, Lipid nanoparticles (LNPs), GalNAc conjugates | Cellular and in vivo nucleic acid delivery | Optimize for cell type-specific uptake and endosomal escape efficiency |
| Splicing Detection | NanoString nCounter, RT-PCR with junction probes, RNA-seq | Quantify alternative splicing isoforms and gene expression | Normalize to constitutive exons, account of PCR amplification biases |
Market Landscape and Future Directions
The RNA therapeutics market demonstrates robust growth, projected to reach USD 22.37 billion by 2032 with a 9.4% CAGR from 2026 [64]. This expansion is driven by technological advances, increased R&D investment, and regulatory familiarity gained during the COVID-19 pandemic [65]. The market is segmented by:
- Therapeutic modality: mRNA (35.7% share), antisense oligonucleotides, siRNA, self-amplifying RNA (22.5% CAGR) [65]
- Application: Oncology (34.2% share, 15.2% CAGR), genetic disorders, infectious diseases [65]
- Geography: North America leads (36.2% share), with Asia Pacific growing rapidly (18.9% CAGR) [65]
Future development will focus on overcoming persistent challenges:
- Delivery optimization: Improving endosomal escape efficiency (currently <10% of LNP cargo reaches cytosol) through novel ionizable lipids and biological vectors [65] [60]
- Extrahepatic targeting: Developing systems to reach neurological, muscular, and tumor tissues beyond the liver [65] [60]
- Combination therapies: Co-delivering multiple RNA therapeutics (e.g., siRNA + mRNA) to target complementary pathways [60]
- Personalized approaches: Leveraging cancer-specific splicing neoantigens for immunotherapy [58]
The trajectory of RNA-targeting therapeutics reflects the evolutionary versatility of RNA itself - from primordial multifunctional molecule to precision medical tool. As delivery technologies advance and our understanding of RNA biology deepens, these therapies will increasingly expand the druggable genome, ultimately fulfilling the therapeutic potential suggested by RNA's foundational role in the origin of life.
The origin of life represents one of science's most profound puzzles, centering on how biological complexity arose from simple prebiotic chemistry. For decades, the RNA world hypothesis has dominated scientific thinking, proposing that self-replicating RNA molecules served as the primordial genetic system before the evolution of DNA and proteins [38]. This hypothesis gains support from RNA's dual capabilities: information storage like DNA and catalytic function like proteins, as evidenced by the discovery of ribozymes [3]. However, this RNA-centric view faces significant challenges, including RNA's chemical instability and the difficulty of explaining how RNA could have emerged spontaneously from prebiotic conditions [66].
A parallel concept, Christian de Duve's "thioester world" hypothesis, proposed that thioesters provided the essential energy source for early metabolic processes before the advent of modern energy carriers like ATP [33]. Thioesters, characterized by a sulfur atom linked to a carbonyl group, are high-energy compounds that drive critical biochemical reactions in extant life, particularly in metabolic pathways like fatty acid synthesis [67]. Recent groundbreaking research has now bridged these two theories, demonstrating that thioesters could have played a fundamental role in activating amino acids for peptide synthesis in an RNA-rich prebiotic environment [28] [33]. This synthesis provides a plausible pathway for the emergence of the first functional peptides alongside replicating RNA systems, offering solutions to key challenges in origins of life research.
The Chemical Principles of Thioester Reactivity
Thioesters are organosulfur compounds with the general structure R-C(=O)-SR'. This structural arrangement confers unique chemical properties that make them particularly valuable in prebiotic chemistry. The key to their reactivity lies in the thioester bond, which is less stable and more reactive than the corresponding oxoester bond found in conventional esters. This relative instability translates to higher free energy, making thioesters effective acyl group carriers and activation agents [67].
The enhanced reactivity of thioesters compared to oxoesters stems from several factors. The carbon-sulfur bond in thioesters is longer and weaker than the carbon-oxygen bond in oxoesters due to poorer p-orbital overlap between carbon and sulfur atoms. Additionally, the sulfur atom stabilizes the adjacent carbanion less effectively than oxygen would, making the carbonyl carbon more electrophilic and susceptible to nucleophilic attack [68]. This property is crucial for peptide bond formation, as the activated carbonyl can be attacked by the amino group of another amino acid.
In modern biochemistry, thioesters remain essential intermediates in numerous metabolic pathways. Most notably, they feature prominently in coenzyme A (CoA) derivatives, such as acetyl-CoA, which serves as a central hub in metabolism [38]. The persistence of thioesters throughout evolutionary history suggests they may represent molecular fossils from life's earliest beginnings, providing a continuous thread from prebiotic chemistry to contemporary biological systems [33].
Thioester-Mediated Peptide Synthesis: Core Mechanism
The central premise of thioester-mediated peptide synthesis involves using thioesters to overcome the significant thermodynamic and kinetic barriers to amide bond formation in aqueous solution. In the absence of activation, amino acids in water favor the formation of protonated or zwitterionic structures that are thermodynamically stable and resistant to condensation. Thioesters provide the necessary activation energy to make peptide bond formation favorable under prebiotic conditions.
The mechanism proceeds through a series of well-defined steps that have been demonstrated to occur spontaneously under plausible early Earth conditions:
Activation via Thioester Formation
The process begins with the conversion of free amino acids into amino acid thioesters. Recent research has shown that this activation can occur using pantetheine, a precursor to coenzyme A that has been synthesized under prebiotic conditions [28] [33]. The thioesterification reaction provides the necessary energy landscape for subsequent condensation.
RNA Aminoacylation
The activated amino acid thioesters then react with RNA molecules, forming aminoacyl-RNA conjugates. This critical step, demonstrated by Powner and colleagues, occurs spontaneously in water at neutral pH and represents the first stage of coupling the genetic and functional worlds [28]. The reaction is notable for its chemical selectivity, favoring attachment to RNA over competing side reactions.
Peptide Bond Formation
Once linked to RNA, the amino acid thioesters become primed for peptide bond formation. The RNA-bound thioesters react with incoming free amino acids or other aminoacyl-RNA conjugates, extending the peptide chain. This process benefits from proximity effects and catalytic assistance potentially provided by the RNA scaffold itself [28].
Table 1: Key Advantages of Thioester-Mediated Peptide Synthesis in Prebiotic Chemistry
| Advantage | Chemical Principle | Prebiotic Significance |
|---|---|---|
| Water Compatibility | Reactions proceed in aqueous environments despite hydrolysis challenges | Plausible in early Earth aquatic environments like pools or lakes |
| Energy Efficiency | Utilizes thioester bond energy without requiring additional activation | Functions without complex modern enzymatic machinery |
| Selective Activation | Activates carboxyl groups without protecting groups | Provides regioselectivity under messy prebiotic conditions |
| RNA Coupling | Spontaneously forms aminoacyl-RNA conjugates | Bridges "RNA world" and "protein world" hypotheses |
| Extended Peptide Synthesis | Supports formation of peptides longer than dimers | Enables synthesis of functionally relevant peptides |
Experimental Protocols and Methodologies
Standard Protocol for Thioester-Mediated RNA Aminoacylation
The following protocol, adapted from Singh et al. (2025), details the experimental procedure for demonstrating spontaneous amino acid attachment to RNA using thioester chemistry [28]:
Reagents and Preparation:
- Prepare amino acid thioesters fresh by reacting free amino acids with pantetheine under mild aqueous conditions. Pantetheine synthesis follows previously established prebiotic pathways using hydrogen cyanide as a starting material [33].
- Use short RNA oligonucleotides (10-15 nucleotides) representing minimalistic prebiotic RNA sequences.
- Employ a neutral pH buffer (pH 7.0-7.5) to simulate plausible early Earth aquatic environments.
- Conduct reactions under inert atmosphere (N₂ or Ar) to minimize oxidation side reactions.
Procedure:
- Reaction Setup: In a 1.5 mL microcentrifuge tube, combine RNA oligonucleotide (0.5 mM final concentration) with amino acid thioester (2.0 mM final concentration) in neutral pH buffer.
- Incubation: Allow the reaction to proceed at 25°C for 24-48 hours without agitation.
- Monitoring: Track reaction progress using analytical HPLC with UV detection at 260 nm (RNA absorbance) and 220 nm (peptide bond formation).
- Verification: Confirm aminoacyl-RNA formation using tandem mass spectrometry (LC-MS/MS) to detect mass addition corresponding to specific amino acids.
- Peptide Extension: To demonstrate peptide synthesis, add free amino acids (5.0 mM final concentration) to the pre-formed aminoacyl-RNA and continue incubation for an additional 24-48 hours.
Key Observations:
- The reaction proceeds efficiently without external catalysis or coupling agents.
- HPLC analysis shows gradual disappearance of starting materials and emergence of new peaks corresponding to aminoacyl-RNA conjugates.
- Mass spectrometry confirms the identity of reaction products through exact mass matching and fragmentation patterns.
- The system demonstrates versatility across multiple amino acid types, though with variations in efficiency [28].
Kinetic Analysis of Thioester Exchange Reactions
For quantitative assessment of thioester reactivity, kinetic studies can be performed using the SEA (bis(2-sulfanylethyl)amido) peptide system [69]:
Experimental Setup:
- Prepare SEA peptide substrate at 1 mM concentration in 6 M guanidine hydrochloride buffer at pH 4.0.
- Use tris(2-carboxyethyl)phosphine (TCEP) as reducing agent (100 mM) to maintain thiol/selenol catalysts in active form.
- Employ thiol additives like 3-mercaptopropionic acid (MPA) at 5% v/v as thioester acceptors.
- Test catalytic efficiency of various selenol catalysts (e.g., compounds 13, 14, and 8a from [69]) across concentration range of 6.25-200 mM.
Data Collection:
- Monitor reaction progress by HPLC with regular sampling over 8-12 hours.
- Calculate apparent second-order rate constants from concentration-time data.
- Determine half-reaction times (t₁/₂) for comparative analysis of catalytic efficiency.
Table 2: Kinetic Parameters for Selenol-Catalyzed Thioester Formation from SEA Peptides
| Catalyst | Concentration (mM) | Half-Reaction Time (h) | Relative Rate Enhancement |
|---|---|---|---|
| Uncatalyzed | - | 7.28 | 1.0x |
| 8a | 6.25 | 3.35 | 2.2x |
| 13 | 6.25 | 5.87 | 1.2x |
| 8a | 50 | 1.95 | 3.7x |
| 13 | 50 | 2.22 | 3.3x |
| 8a | 200 | 1.97 | 3.7x |
| 13 | 200 | 1.68 | 4.3x |
Data adapted from [69]
Integration with the RNA World Hypothesis
The discovery of thioester-mediated peptide synthesis provides critical missing links in the RNA world hypothesis, addressing several fundamental limitations of RNA-only scenarios. The spontaneous coupling of amino acids to RNA molecules offers a natural mechanism for the emergence of the first genetic code, where specific RNA sequences could potentially selectively bind particular amino acids based on structural compatibility [28]. This represents a plausible initial step toward the sequence-specific peptide synthesis that characterizes modern biology.
This integration also helps resolve the catalytic limitations of the RNA world. While ribozymes demonstrate impressive catalytic capabilities, their functional diversity and efficiency pale in comparison to protein enzymes. The early association of peptides with RNA could have provided immediate functional benefits, with peptides serving as cofactors enhancing or expanding ribozyme capabilities [38]. This synergistic relationship might have been a crucial evolutionary step toward the protein-dominated catalysis of contemporary biology.
Furthermore, thioester chemistry bridges the gap between early metabolism and replication. The energy-carrying capacity of thioesters aligns with de Duve's "thioester world" concept, suggesting they could have powered both metabolic reactions and the synthesis of information-containing polymers [33]. This dual functionality positions thioesters as central players in a unified origins scenario where replication and metabolism co-evolved rather than emerging separately.
Contemporary Applications and Research Tools
The principles of thioester-mediated activation have found important applications in modern chemical biology and pharmaceutical development. Beyond their origins relevance, thioesters serve as crucial intermediates in peptide synthesis and protein engineering, particularly in Native Chemical Ligation (NCL) strategies for protein semi-synthesis [69] [70]. The development of selenol-based catalysts for thiol-thioester exchange reactions has improved the efficiency of these processes, enabling more effective production of peptide thioesters for protein chemical synthesis [69].
Recent innovations include enzymatic approaches for thioester generation. A 2025 study described an engineered bacterial E1-like enzyme that enables ATP-driven activation of protein and peptide C-termini for thioester formation [70]. This methodology provides high-yield, specific generation of thioesters for bioconjugation applications, addressing previous limitations in C-terminal modification strategies.
In biochemical research, thioesters have been successfully incorporated into modern translation systems. Remarkably, studies have shown that 3'-thio-tRNAs, where the native oxoester is replaced by a thioester, are effectively recognized and utilized by wild-type E. coli ribosomes [68]. This backward compatibility suggests that modern protein synthesis machinery may have evolved from simpler thioester-dependent systems, preserving recognition capabilities for these primordial chemistry relics.
Research Reagent Solutions
Table 3: Essential Research Reagents for Thioester-Mediated Peptide Synthesis Studies
| Reagent/Catalyst | Chemical Function | Application in Research |
|---|---|---|
| Pantetheine | Thiol compound for thioester formation | Prebiotic amino acid activation; coenzyme A precursor |
| Selenol Catalyst 13 | Low pKa selenol for catalysis | Accelerates thiol-thioester exchange at mildly acidic pH |
| Selenol Catalyst 8a | Bis(selenol) tertiary amine compound | High-efficiency catalysis of SEA/thiol exchange reactions |
| MPA (3-Mercaptopropionic Acid) | Thiol additive | Classical thiol for preparing peptide thioesters |
| TCEP | Reducing agent | Maintains thiol/selenol catalysts in active reduced state |
| SEA Peptide System | N,S-acyl shift system | Model for studying thioester formation from peptides |
Thioester-mediated activation represents a compelling mechanism for peptide synthesis in prebiotic environments, effectively bridging the gap between the "RNA world" and "protein world" hypotheses. The demonstrated capacity of thioesters to spontaneously link amino acids with RNA under plausible early Earth conditions provides a chemically robust pathway for the emergence of the first functional peptides alongside replicating genetic elements. This synergistic relationship addresses critical limitations of RNA-only scenarios while preserving the central role of RNA in early evolution.
Significant challenges and opportunities remain for future research. The sequence specificity of RNA-amino acid interactions represents a crucial next frontier—understanding how specific RNA sequences might preferentially bind particular amino acids could illuminate the origins of the genetic code [28]. Additionally, exploring the potential for ribozyme-catalyzed thioester formation and peptide bond formation would further strengthen the integrated model. From a practical perspective, optimizing thioester chemistry for pharmaceutical applications, particularly in peptide therapeutic synthesis and protein engineering, continues to be an active area of innovation [67] [70].
The resurrection of thioester chemistry as a central theme in origins of life research demonstrates how investigating life's beginnings continues to yield valuable insights and tools for contemporary science. As research progresses, the integration of thioester-mediated activation with other prebiotic synthesis pathways will likely provide an increasingly coherent picture of life's chemical origins, while simultaneously inspiring new methodologies for synthetic biology and chemical synthesis.
The RNA World hypothesis posits that RNA was the primordial biopolymer, serving as both a catalyst for chemical reactions and a store of genetic information before the evolutionary emergence of DNA and proteins [71] [38]. This central role of RNA in life's origin is evidenced by its dual capabilities: like DNA, it can store and replicate genetic information, and as a ribozyme, it can catalyze critical chemical reactions [38]. The ribosome, the core machinery for protein synthesis, is itself a ribozyme, strengthening the hypothesis that an RNA-based life form preceded our current DNA/RNA/protein system [3] [71]. Today, this ancient molecule is spearheading a modern therapeutic revolution. Messenger RNA (mRNA) technology has emerged as a versatile platform capable of preventing infectious diseases, treating cancers, and replacing deficient proteins, thereby translating fundamental research on prebiotic chemistry into a new pillar of modern medicine [72] [73].
This whitepaper provides an in-depth technical guide to mRNA therapeutic platforms. It examines the core technology, delivery systems, analytical methods, and the expanding clinical applications that are reshaping drug development.
Core mRNA Technology and Delivery Mechanisms
Engineering the mRNA Molecule for Therapeutic Use
A therapeutic mRNA molecule is a sophisticatedly engineered construct designed for high stability, minimal immunogenicity, and efficient translation in vivo [72] [74]. Its architecture comprises several critical elements:
- 5' Cap: A modified guanine nucleotide essential for ribosome binding, translation initiation, and protecting the mRNA from exonuclease degradation [72] [75].
- 5' and 3' Untranslated Regions (UTRs): These flanking regions regulate mRNA stability, localization, and translational efficiency [76] [74].
- Open Reading Frame (ORF): The coding sequence for the desired therapeutic protein. Codon optimization is often employed to enhance translational efficiency and yield [72].
- Poly(A) Tail: A sequence of adenine nucleotides at the 3' end that significantly enhances mRNA stability and translation [72] [75].
Key advancements that enabled the clinical success of mRNA therapeutics include the incorporation of chemically modified nucleosides (e.g., pseudouridine, 5-methylcytidine) and extensive sequence engineering. These modifications reduce the innate immunogenicity of the mRNA and increase its stability and translational capacity [76] [74].
Lipid Nanoparticles: The Essential Delivery Vehicle
The delivery of fragile mRNA molecules into the cytoplasm is achieved primarily via Lipid Nanoparticles (LNPs) [72] [76]. LNPs are multi-component systems that protect mRNA and facilitate its cellular uptake and endosomal escape. The table below details the standard components of an LNP formulation.
Table: Key Components of Lipid Nanoparticles (LNPs) for mRNA Delivery
| Component | Function | Examples/Chemical Classes |
|---|---|---|
| Ionizable Cationic Lipid | Essential for mRNA encapsulation and endosomal escape; positively charged at low pH to interact with endosomal membrane. | DLin-MC3-DMA, SM-102, ALC-0315 [76] |
| Phospholipid | Structural lipid that forms the LNP bilayer and supports fusion with cellular membranes. | DSPC [76] |
| Cholesterol | Stabilizes the LNP structure and enhances integrity and cellular uptake. | Cholesterol [76] |
| PEGylated Lipid | Shields the LNP surface, reduces non-specific binding, controls particle size, and improves stability in circulation. | DMG-PEG 2000 [72] [76] |
Recent innovations in LNP technology focus on overcoming the natural hepatotropism (liver-targeting) of first-generation LNPs. Strategies for tissue-specific mRNA delivery include:
- SORT Technology: The addition of supplemental, charge-altering "SORT" molecules (cationic, anionic, or ionizable lipids) to traditional LNPs to redirect mRNA translation to specific organs like the spleen or lungs [76].
- Ligand Conjugation: Functionalizing the LNP surface with antibodies, peptides, or other targeting moieties to enhance cell-type-specific tropism, e.g., targeting T cells or gut-homing leukocytes [76].
- High-Throughput Screening: Using DNA-barcoded LNP libraries to screen thousands of lipid structures in parallel for their ability to target specific cell types in vivo [76].
Diagram: Mechanism of Action of mRNA-LNP Therapeutics
Analytical Strategies for mRNA Therapeutic Characterization
Ensuring the safety, efficacy, and quality of mRNA therapeutics requires a comprehensive analytical strategy to characterize critical quality attributes (CQAs). The following workflow outlines the key analytical steps from initial synthesis to final product release.
Diagram: mRNA Analytical Characterization Workflow
The table below summarizes the primary analytical techniques employed to assess these CQAs, providing a toolkit for research and quality control (QC).
Table: Essential Analytical Techniques for mRNA Therapeutic Characterization
| Quality Attribute | Analytical Technique | Technical Summary & Purpose |
|---|---|---|
| Integrity/Purity | Capillary Gel Electrophoresis (CGE) | High-resolution separation based on size-to-charge ratio to quantify full-length mRNA and degradants [75]. |
| Ion-Pair Reversed-Phase HPLC (IP-RP HPLC) | Separates mRNA from impurities (abortive transcripts, dsRNA) based on hydrophobic interactions [75]. | |
| Identity/Sequence | RT-PCR & Sanger Sequencing | Conventional method for confirming the Open Reading Frame (ORF) sequence [75]. |
| LC-MS/MS Oligonucleotide Mapping | Provides detailed sequence confirmation and identification of chemical modifications [75]. | |
| Capping Efficiency | HPLC with UV/MS Detection | Quantifies the percentage of mRNA molecules possessing the 5' cap structure, crucial for translation [75]. |
| Poly(A) Tail Length | High-Resolution Gel Electrophoresis | Assesses the length distribution of the poly(A) tail, a key factor in mRNA stability [75]. |
| Impurities (dsRNA) | ELISA or Gel Electrophoresis | Detects and quantifies double-stranded RNA (dsRNA) impurities, which can trigger unwanted innate immune responses [75]. |
| Functionality | In Vitro Translation Assay | Confirms the mRNA's ability to be translated into the full-length, functional protein in a cell-free system [75]. |
| Cell-Based Assays / Western Blot | Assesses biological activity and protein expression in a relevant cellular context [75]. |
Clinical Applications and Commercial Landscape
A Expanding Universe of Therapeutic Indications
Therapeutic mRNA applications are broadly classified into four main categories, demonstrating the platform's remarkable versatility [73].
Table: Classification of mRNA Therapeutics in Clinical Development
| Therapeutic Category | Subcategories & Examples | Mechanism of Action |
|---|---|---|
| Vaccines | - Viral Antigens: COVID-19, Zika, Influenza- Cancer Antigens: Personalized neoantigens, Tumor-Associated Antigens (TAAs)- Bacterial/Antigens: Under investigation [72] [73] | mRNA encoding a pathogen or tumor antigen is delivered to Antigen Presenting Cells (APCs), leading to cellular and humoral immune activation [72]. |
| Protein Replacement | - Maintenance Therapeutics: For genetic disorders (e.g., missing enzymes)- Interventional Therapeutics: VEGF-A for ischemic heart disease, cytokines for cancer [73] [74] | mRNA encoding a functional protein is delivered to cells, transiently replacing a deficient or missing protein, restoring physiological function [74]. |
| Therapeutic Antibodies | mRNA sequences encoding the heavy and light chains of clinically relevant antibodies [73] | The patient's own cells become biofactories, producing therapeutic antibodies in vivo, bypassing complex recombinant protein manufacturing [73]. |
| Cell & Gene Therapy | - mRNA-mediated CRISPR/Cas9- In vivo generation of CAR-T cells [73] | mRNA enables transient, efficient expression of gene-editing tools (e.g., Cas9) or chimeric antigen receptors (CARs) directly in vivo for precise therapeutic interventions [73] [76]. |
Market Outlook and Key Industry Players
The global market for mRNA therapeutics is in a dynamic state of evolution. Following the historic peak driven by COVID-19 vaccines, the market is projected to consolidate and then grow significantly as new applications mature, with one report forecasting a rise from USD 52.59 billion in 2025 to USD 257.11 billion by 2032, representing a compound annual growth rate (CAGR) of 25.41% [77]. Another analysis values the market at $7.71 billion in 2025, expecting stabilization before the next wave of innovations [78].
The field is led by several key players, each with a diversified pipeline:
- Moderna, Inc.: A leader with a broad portfolio spanning infectious diseases, oncology, cardiovascular diseases, and rare diseases [78].
- BioNTech SE: Known for its COVID-19 vaccine, the company has a deep focus on cancer immunotherapies and personalized oncology vaccines [78].
- CureVac N.V.: A pioneer in RNA technology, now partnered with GSK, focusing on next-generation vaccines and cancer therapies [78].
The Scientist's Toolkit: Key Reagents for mRNA Production
The production of high-quality IVT mRNA requires a suite of specialized reagents and materials. The following table details the essential components of a standard mRNA synthesis reaction and their functions.
Table: Essential Research Reagents for In Vitro Transcription (IVT) mRNA Synthesis
| Reagent / Material | Function in the Experimental Protocol |
|---|---|
| Linearized DNA Plasmid Template | Serves as the template from which mRNA is transcribed. The plasmid must be linearized downstream of the poly(A) tail sequence to ensure precise transcription termination [74]. |
| DNA-Dependent RNA Polymerase | The enzyme that catalyzes the synthesis of mRNA from the DNA template. T7, T3, and SP6 polymerases are commonly used [74]. |
| Nucleoside Triphosphates (NTPs) | The building blocks (ATP, UTP, GTP, CTP) for the nascent mRNA strand. Chemically modified NTPs (e.g., N1-methylpseudouridine) are often used to enhance stability and reduce immunogenicity [76] [74]. |
| 5' Cap Analog | Incorporated during or after transcription to form the 5' cap structure, which is essential for translation initiation and stability (e.g., CleanCap) [75] [74]. |
| Reaction Buffer (with Mg²⁺) | Provides the optimal ionic strength and pH (e.g., HEPES/Tris) and supplies magnesium ions, a critical cofactor for polymerase activity [74]. |
| RNase Inhibitors | Protects the fragile mRNA product from degradation by ribonucleases during the synthesis and purification processes [74]. |
| Pyrophosphatase | Degrades inorganic pyrophosphate, a byproduct of the polymerization reaction, which can inhibit the transcription process and lead to premature termination [74]. |
mRNA technology represents a paradigm shift in therapeutics, echoing the functional versatility of RNA from the ancient RNA World. From its foundational success in vaccines, the platform is rapidly expanding into protein replacement, in vivo antibody production, and gene editing. While challenges in targeted delivery and manufacturing robustness remain, continuous innovations in LNP technology, mRNA engineering, and analytical science are paving the way for a new era of precision medicine. The ongoing clinical trials and robust pipelines of industry leaders signal that mRNA platforms will remain a transformative force in global healthcare for decades to come.
Overcoming Prebiotic Challenges: Fidelity, Stability, and Environmental Constraints
Solving the Strand Separation Problem in Early RNA Replication
The RNA World Hypothesis posits that RNA once served as both the primary genetic material and catalytic molecule in early life forms [4] [3]. A critical requirement for Darwinian evolution in such a world is the ability of RNA to self-replicate. However, a significant obstacle to non-enzymatic replication is the strand separation problem: after a template-directed synthesis, the newly formed complementary strands form a stable duplex that does not readily dissociate for subsequent replication cycles [79]. Without a mechanism for strand separation, the replication process grinds to a halt after a single round. This article examines prebiotically plausible solutions to this fundamental problem, focusing on pH-driven denaturation and invader-mediated strand displacement, and their implications for the feasibility of the RNA World.
Prebiotically Plausible Strand Separation Mechanisms
pH-Driven Strand Separation
Geological environments on early Earth likely experienced natural pH fluctuations. Research has demonstrated that these oscillations could have been harnessed to drive RNA strand separation under moderate temperatures, avoiding the high-temperature degradation associated with thermal denaturation [80].
Key Experimental Findings:
- Reduced Melting Temperature: Changes in pH can significantly tune the melting temperature (Tm) of oligoribonucleotide duplexes, enabling strand separation at temperatures as low as 0°C under acidic conditions, compared to over 40°C required at neutral pH [80].
- Enhanced RNA Stability: Acid denaturation conditions reduce the risk of phosphodiester bond cleavage, a major degradation pathway for RNA, both in the presence and absence of divalent metal ions [80].
- Plausible Geological Context: The process aligns with plausible geochemical scenarios, such as pH oscillations in volcanic settings or hydrothermal vents, providing a natural mechanism for achieving replication cycles [80].
Invader-Mediated Strand Displacement
Inspired by biological processes and DNA nanotechnology, studies have explored non-enzymatic strand displacement using short "invader" oligonucleotides. This mechanism does not require bulk environmental changes and could operate within a protocellular compartment [79].
Mechanism of Action:
The process utilizes a toehold/branch migration mechanism. A short invader strand first binds to a single-stranded toehold region (an overhang) on a "blocker" strand that is complementary to the template. The invader then fully displaces the blocker from the template through branch migration, freeing the template for a new round of primer extension [79].
Key Experimental Parameters:
Optimization studies revealed that reaction efficiency is highly dependent on invader length and concentration, as well as temperature. For example, an 8-nucleotide invader achieved a maximal primer extension rate of 0.9 ± 0.1 h⁻¹ at room temperature, while a 6-nucleotide invader required lower temperatures to reach similar efficiency, likely due to enhanced toehold binding stability [79].
Comparative Analysis of Strand Separation Mechanisms
Table 1: Comparison of Strand Separation Mechanisms for Early RNA Replication
| Feature | pH-Driven Denaturation | Invader-Mediated Displacement |
|---|---|---|
| Core Principle | Modifies protonation of nucleobases to destabilize duplex [80] | Uses toehold binding and branch migration to physically displace strand [79] |
| Environmental Requirement | Oscillating pH (e.g., acidic for separation, neutral for copying) [80] | Supply of short oligonucleotide invaders [79] |
| Prebiotic Plausibility | High; compatible with volcanic or hydrothermal settings [80] | Moderate; requires a source of short, specific RNA oligomers [79] |
| Compatibility with Protocells | Challenging; bulk pH changes may be difficult to confine [80] | High; can operate within a confined compartment [79] |
| Key Advantage | Avoids high temperatures and reduces RNA degradation [80] | Enables continuous, enzyme-free replication without bulk environmental swings [79] |
| Primary Challenge | Achieving rapid and localized pH cycling [80] | Ensuring efficient invasion against stable duplexes [79] |
Detailed Experimental Protocols
Protocol for Studying pH-Driven Strand Separation
This protocol is adapted from studies investigating RNA duplex stability under varying pH conditions [80].
1. Sample Preparation:
- RNA Duplexes: Synthesize and anneal complementary oligoribonucleotides to form stable duplexes.
- Buffer Systems: Prepare a range of buffers covering pH 3.0 to 8.0. Citrate-phosphate buffers are suitable for acidic conditions, while phosphate buffers are used for neutral pH.
2. Melting Temperature (Tm) Analysis:
- Method: Use Ultraviolet (UV) spectrophotometry to monitor absorbance at 260 nm while applying a controlled temperature gradient.
- Data Collection: Record absorbance changes to determine the melting temperature (Tm) at each pH value. The Tm is defined as the temperature at which 50% of the duplex is dissociated.
- Analysis: Plot Tm versus pH to quantify the effect of pH on duplex stability.
3. Strand Separation and Replication Assessment:
- Separation Confirmation: Use gel electrophoresis under non-denaturing conditions to visualize strand separation after exposure to low pH and subsequent neutralization.
- Replication Compatibility: Test the functionality of the separated strands as templates for non-enzymatic primer extension reactions using activated nucleotides (e.g., 5'-5'-imidazolium-bridged dinucleotides) at neutral pH [79].
Protocol for Non-Enzymatic Primer Extension with Strand Displacement
This protocol details the methodology for demonstrating invader-facilitated RNA synthesis [79].
1. RNA Complex Assembly:
- Prepare a primer/template duplex.
- Hybridize a full-length "blocker" strand to the template, ensuring it includes a 5' or 3' single-stranded toehold region (typically 6-8 nucleotides).
2. Strand Displacement Reaction:
- Reaction Mixture: Combine the pre-assembled primer/template/blocker complex with the invader oligonucleotide and the activated nucleotide substrate (e.g., Imidazolium-bridged C*C for templating GG).
- Key Variables:
- Invader Concentration: Test a range from sub-stoichiometric to saturating levels (e.g., 1-100 µM).
- Divalent Ions: Include Mg²⁺ (e.g., 0-100 mM), which catalyzes the primer extension step.
- Temperature: Conduct reactions at different temperatures (e.g., 0°C, 25°C) to optimize invader binding versus reaction rate.
3. Product Analysis:
- Use denaturing polyacrylamide gel electrophoresis (PAGE) to separate and visualize the reaction products.
- Quantify the extent of primer extension by measuring the conversion of the primer to longer products. Calculate pseudo-first-order reaction rates (kobs) from the time-dependent disappearance of the unreacted primer.
Signaling Pathways and Workflow Visualizations
Comparative Strand Separation Pathways
Invader-Mediated Strand Displacement Workflow
The Scientist's Toolkit: Key Research Reagents
Table 2: Essential Reagents for Studying Non-Enzymatic RNA Replication
| Reagent / Material | Function in Experiment | Technical Notes |
|---|---|---|
| Oligoribonucleotides | Serve as templates, primers, blockers, and invaders. Fundamental building blocks for replication assays. | Requires chemical synthesis. Sequence design is critical, especially for toehold length (e.g., 6-8 nt) and invader complementarity [79]. |
| Activated Nucleotides | Substrates for non-enzymatic template-directed RNA synthesis. Provide the chemical energy for phosphodiester bond formation. | 2-aminoimidazole-activated monomers or 5'-5'-imidazolium-bridged dinucleotides are common prebiotically plausible choices [79]. |
| pH Buffer Systems | Create and maintain specific pH environments to study pH-dependent strand separation and reaction kinetics. | Citrate-phosphate for acidic pH; phosphate or other buffers for neutral pH. Must be compatible with RNA integrity [80]. |
| Divalent Metal Ions (e.g., Mg²⁺) | Catalyze the chemical step of primer extension. Can also influence RNA duplex stability and strand displacement efficiency. | Concentration must be optimized; high levels can promote RNA degradation [79]. |
| Denaturing PAGE Gels | Analyze reaction products, separate extended primers from starting materials, and quantify replication efficiency. | Standard method for resolving small RNA fragments. Provides high resolution for products differing by single nucleotides. |
The strand separation problem represents a major hurdle for the RNA World Hypothesis. The experimental approaches detailed here—pH-driven denaturation and invader-mediated strand displacement—demonstrate that non-enzymatic, prebiotically plausible solutions to this problem are feasible. pH oscillations leverage likely geological conditions to gently separate strands, while molecular displacement mechanisms mimic modern biological strategies. These findings significantly strengthen the case for a replicative RNA world by providing tangible pathways for the iterative cycles of replication necessary for Darwinian evolution. Future research will focus on integrating these separation mechanisms with continuous, genome-length replication within model protocells, bringing us closer to recapitulating the origins of life in the laboratory.
The RNA world hypothesis posits a stage in the early evolution of life where RNA molecules served both as the genetic material and the primary catalytic agents, preceding the advent of DNA and proteins [81]. A critical challenge for origin-of-life studies is to understand how a protein-free RNA world could have become established on the primitive Earth, given the intrinsic chemical instabilities and synthetic difficulties associated with RNA polymers [11] [82]. This in-depth guide explores how specific environmental conditions—pH, temperature cycles, and mineral catalysis—could have optimized prebiotic chemistry to overcome these hurdles. Environmental parameters are not merely a backdrop but are hypothesized to be active, enabling factors that selected for the first self-replicating molecular systems. By examining the interplay between these factors and prebiotic chemistry, this guide provides a technical framework for experimental approaches to the RNA world, aimed at researchers and scientists investigating the origins of life.
The Role of pH in Prebiotic System Stability and Function
The pH of the primordial environment is a critical factor influencing the stability, structure, and catalytic activity of prebiotic molecules, particularly RNA. While RNA is notoriously susceptible to base-catalyzed hydrolysis at neutral to alkaline pH ( >6) [82], recent research suggests that moderately acidic conditions (pH 4–5) offer several distinct advantages for the emergence of an RNA world.
Enhanced Molecular Stability at Acidic pH
At acidic pH, the phosphodiester bonds of the RNA backbone and the ester bonds critical for activating monomers for polymerization demonstrate significantly greater stability [82]. This directly counters one of the principal objections to the RNA world—the inherent instability of the RNA molecule. Furthermore, protonation of nucleotide bases under these conditions can open new structural and functional possibilities.
- The RNA i-Motif: A key proposal is that cytosine residues, which are particularly unstable and prone to deamination, could have been stabilized in the RNA i-motif structure [83]. This four-stranded quadruplex is formed by cytosine-rich sequences under acidic conditions (pH ~6.5 or lower), where protonated (C+) and unprotonated (C) cytosine residues form stable base pairs [83]. This structure not only protects cytosine from degradation by slowing its deamination rate but also provides a potential scaffold for binding ligands and performing early functional roles [83].
- Reduced Dependence on Divalent Cations: The folding and catalytic activity of many ribozymes require divalent metal ions like Mg²⁺. However, high concentrations of Mg²⁺ can also catalyze RNA degradation, creating a paradoxical problem for template copying in a prebiotic setting [82]. Acidic conditions can mitigate this issue. For instance, a self-cleaving ribozyme with maximum activity at pH 4 was found to be active in the complete absence of divalent ions, suggesting that protonated nucleotides could fulfill some of the structural roles typically played by metal ions [82].
Table 1: Impact of pH on Key Prebiotic Molecules and Processes
| Parameter | Alkaline/Near-Neutral Conditions (pH >7) | Acidic Conditions (pH 4–6) | Experimental Evidence |
|---|---|---|---|
| RNA Backbone Stability | Low; susceptible to base-catalyzed hydrolysis [82] | High; phosphodiester bonds are more stable [82] | Kinetic studies of RNA degradation rates |
| Cytosine Stability | Low; rapid deamination [83] | High; stabilized in i-motif structures [83] | Measurement of deamination rates in i-motifs |
| Metal Ion Requirement | High; Mg²⁺ often essential for folding and catalysis [82] | Reduced; some ribozymes active without Mg²⁺ [82] | In vitro selection and activity assays of ribozymes |
| Prebiotic "Soup" Analogy | Dilute aqueous solution | Vinaigrette or mayonnaise (emulsified) [82] | — |
Experimental Protocols for Investigating pH Effects
Protocol 1: Quantifying RNA Stability Under Variable pH
- Objective: To determine the half-life of RNA oligonucleotides under different pH conditions.
- Methodology:
- Sample Preparation: Prepare identical samples of a specific RNA sequence (e.g., a short oligoribonucleotide) in buffered solutions across a pH range (e.g., 4.0, 5.0, 6.0, 7.0, 8.0).
- Incubation: Incubate all samples at a constant, relevant temperature (e.g., 25°C or 40°C). Aliquots will be removed at regular time intervals.
- Analysis: Analyze aliquots via denaturing polyacrylamide gel electrophoresis (PAGE) to separate intact RNA from degradation products. The RNA can be visualized using stains like SYBR Gold.
- Quantification: Use densitometry to quantify the amount of full-length RNA remaining at each time point. Plot the natural logarithm of RNA concentration versus time to determine the rate constant (k) and half-life (t₁/₂ = ln(2)/k) for each pH condition.
Protocol 2: Probing i-Motif Formation
- Objective: To confirm the formation of RNA i-motif structures under acidic conditions.
- Methodology:
- Sample Design: Synthesize a cytosine-rich RNA sequence known to form i-motifs.
- Spectroscopic Analysis: Use UV-Vis Spectroscopy to monitor the absorbance at ~295 nm, which is characteristic of i-motif formation, as a function of decreasing pH.
- Structural Confirmation: Employ Circular Dichroism (CD) Spectroscopy. i-Motifs exhibit a distinctive CD spectrum with a positive peak around 285 nm and a negative peak around 260 nm. A titration from neutral to acidic pH will show the emergence of this signature.
- Kinetics: The same CD or UV-Vis setup can be used to measure the rate of cytosine deamination within the i-motif versus in a single-stranded control, testing the hypothesis that the structure confers stability.
Diagram 1: Experimental workflow for RNA stability analysis.
Temperature and Cyclic Conditions for Prebiotic Activation
Temperature is another fundamental environmental variable. While high temperatures can accelerate chemical reactions, they also promote the degradation of complex molecules like RNA. Conversely, low temperatures can stabilize molecules but slow down reaction kinetics. The resolution to this paradox may lie in cyclic temperature variations, which can drive polymerization and selection processes.
The Eutectic Phase and Cold Activation
There is a growing body of evidence supporting the idea that the RNA world may have evolved in icy environments. When aqueous solutions freeze, solutes and molecules become concentrated in a network of liquid veins between ice crystals, a state known as the eutectic phase [82]. This concentration effect can dramatically enhance the rate of polymerization reactions by bringing monomers into close proximity.
- Enhanced Polymerization: Non-enzymatic template-directed polymerization of activated nucleotides proceeds more efficiently in the eutectic phase of ice than in liquid water at room temperature [82].
- Ribozyme Activity at Sub-zero Temperatures: Remarkably, some ribozymes exhibit maximal catalytic activity at temperatures of -7°C to -8°C [82]. This is attributed to the combined effects of increased RNA concentration and lowered water activity within the icy matrix, which can stabilize the active folded structure of the ribozyme.
Experimental Protocols for Simulating Temperature Cycles
Protocol 3: Eutectic-Phase Polymerization
- Objective: To demonstrate the enhancement of non-enzymatic RNA oligomerization in ice.
- Methodology:
- Reaction Setup: Prepare a solution containing activated nucleotides (e.g., ImpN) and a potential template or catalyst (e.g., montmorillonite clay). Use a neutral pH buffer to avoid freezing point depression from strong acids/bases.
- Cycling: Subject the reaction mixture to freeze-thaw cycles (e.g., -20°C for 12 hours, followed by 4°C for 12 hours). Control reactions are kept constantly at 4°C and -20°C.
- Analysis: After a set number of cycles, thaw the samples and analyze the products using ion-exchange chromatography or PAGE to separate and quantify the formed oligomers.
- Comparison: Compare the length and yield of oligomers from the cycled samples versus the static controls.
Protocol 4: Testing Ribozyme Function in Ice
- Objective: To measure the kinetic parameters of a ribozyme (e.g., a self-cleaving hammerhead) at sub-zero temperatures.
- Methodology:
- Sample Preparation: Incorporate the ribozyme and its substrate in a buffered solution. The buffer must be chosen to prevent freezing at the target temperatures if a fully aqueous phase is desired, or the experiment can be set up in a eutectic ice phase.
- Activity Assay: Incubate the reaction at a series of low temperatures (e.g., 0°C, -5°C, -10°C). At timed intervals, quench aliquots with an EDTA-containing stop solution.
- Product Quantification: Analyze the quenched aliquots by PAGE to separate the cleaved product from the uncleaved substrate.
- Kinetic Analysis: Plot the formation of product over time to determine the reaction rate (k_obs) at each temperature, revealing the temperature optimum for activity.
Table 2: Impact of Temperature Regimes on Prebiotic Processes
| Temperature Regime | Advantages | Limitations/Challenges | Key Experimental Findings |
|---|---|---|---|
| Constant Warm (~40-90°C) | Accelerates reaction rates. | Increases RNA hydrolysis and degradation [82]. | Limited success in sustained polymerization. |
| Constant Cold (<0°C) | Enhances stability of RNA and other organics; concentrates reactants in eutectic phase [82]. | Slows down most chemical reaction rates. | Maximal ribozyme activity observed at -7°C to -8°C [82]. |
| Freeze-Thaw Cycles | Combines concentration effect of freezing with reaction acceleration during liquid phases. | Can lead to sequence-dependent mismatches and inactive complexes at low T [82]. | Efficient formation of long RNA oligomers from activated monomers [82]. |
Mineral Catalysis in Monomer Activation and Polymerization
Minerals represent the most primitive and plausible prebiotic catalysts, providing surfaces that could adsorb, concentrate, orient, and catalyze the reactions of organic molecules. Their role is considered indispensable for moving from simple prebiotic precursors to functional biopolymers.
Mechanisms of Mineral Catalysis
Minerals can facilitate prebiotic chemistry through several mechanisms:
- Surface Adsorption and Concentration: Mineral surfaces can adsorb organic molecules from dilute prebiotic solutions, effectively concentrating them and increasing the probability of reaction. This is a fundamental first step for polymerization.
- Templating and Orientation: Certain mineral surfaces possess crystalline structures that can template the formation of polymers. A prime example is the clay mineral montmorillonite, which not only binds activated nucleotides but also catalyzes the formation of predominantly 3',5'-linked RNA-like oligomers [84]. This regiospecificity is critical, as uncatalyzed reactions in solution favor the formation of 2',5'-linkages, which are less compatible with the stable double-helix formation of modern RNA [84].
- Lewis Acid Catalysis: Metal ions within minerals can act as Lewis acids, coordinating to the electronegative oxygen atoms of phosphate groups or carbonyl groups in intermediates. This polarization weakens bonds and makes them more susceptible to nucleophilic attack. For instance, Pb²⁺ and Zn²⁺ have been shown to catalyze the oligomerization of nucleotides [84].
Experimental Protocols for Mineral Catalysis
Protocol 5: Clay-Catalyzed Oligomerization of Nucleotides
- Objective: To demonstrate the mineral-catalyzed formation of RNA oligomers from activated nucleotides.
- Methodology:
- Catalyst Preparation: Purify and characterize montmorillonite clay. The catalytic efficiency can vary significantly between different clay samples [84].
- Reaction Setup: Incubate an activated nucleotide (e.g., adenosine 5'-phosphorimidazolide, ImpA) with the montmorillonite catalyst in a suitable aqueous buffer at neutral pH.
- Product Analysis: After a set period, separate the liquid phase from the clay by centrifugation. Analyze the supernatant using HPLC (High-Performance Liquid Chromatography) to separate and quantify the oligomers based on length.
- Linkage Analysis: To confirm the linkage type (3',5' vs. 2',5'), the oligomers can be digested with specific ribonucleases (e.g., RNase T1, which is specific for 3',5' linkages) and the digestion products analyzed again.
Protocol 6: Testing Metal Ion Catalysis in Sugar-Phosphate Reactions
- Objective: To investigate the catalytic effect of Fe²⁺ and other metal ions on prebiotic reaction networks.
- Methodology:
- Network Simulation: Set up reactions between simple sugars and phosphate sources in the presence of different metal ions (Fe²⁺, Fe³⁺, Mg²⁺, Zn²⁺).
- Product Screening: Use techniques like LC-MS (Liquid Chromatography-Mass Spectrometry) to identify and quantify the range of products formed.
- Kinetic Profiling: Monitor the consumption of reactants and the formation of key products (e.g., ribose-5-phosphate) over time to determine rate enhancements and selectivity induced by the metal catalysts, as demonstrated in studies expanding sugar-phosphate networks [85].
Diagram 2: Multistep mechanism of mineral catalysis.
The Scientist's Toolkit: Essential Research Reagents and Materials
This section details key reagents, materials, and instruments essential for conducting experimental research in prebiotic environmental optimization.
Table 3: Research Reagent Solutions for Prebiotic Chemistry Studies
| Reagent/Material | Specifications & Purity | Primary Function in Experiments |
|---|---|---|
| Activated Nucleotides | e.g., Nucleoside 5'-Phosphorimidazolides (ImpN). >95% purity recommended. | Substrates for non-enzymatic polymerization studies; more reactive than nucleotides for oligomer formation [84]. |
| Montmorillonite Clay | K10 or similar, source-specific. Purified to remove soluble contaminants. | Catalyzes regiospecific (3',5'-linked) oligomerization of RNA monomers; also acts as a substrate concentrator [84]. |
| Buffer Systems | e.g., MES (pKa ~6.1), HEPES (pKa ~7.5). Use of pH buffers tailored to the specific range of interest is critical. | Maintains precise pH conditions to study stability, structure (e.g., i-motif), and reaction rates. |
| Divalent Metal Salts | MgCl₂, FeCl₂, ZnCl₂, Pb(NO₃)₂. High-purity, molecular biology grade. | Investigate metal ion catalysis in polymerization (Pb²⁺) or ribozyme folding/function (Mg²⁺). Fe²⁺ can expand sugar-phosphate networks [85] [84]. |
| RNA Oligonucleotides | Synthetic, HPLC-purified. Specific sequences for i-motif studies (C-rich) or ribozyme cores. | Substrates for stability assays, structural studies (CD, UV), and functional activity tests. |
| Analytical Instruments | HPLC/UPLC, PAGE equipment, UV-Vis Spectrophotometer, Circular Dichroism (CD) Spectrometer. | For separation, quantification, and structural analysis of reactants and products. |
The journey to understand the origin of the RNA world is increasingly focusing on the specific environmental conditions that could have made it possible. The experimental data synthesized in this guide strongly suggest that a unique combination of moderately acidic pH, cyclic low-temperature regimes, and specific mineral catalysts could have collectively solved the fundamental problems of RNA instability, inefficient polymerization, and lack of regiospecificity. Acidic conditions stabilize the molecule and enable novel structures; temperature cycles concentrate reactants and can enhance catalytic function; and mineral surfaces provide the essential scaffolding and catalytic power for forming the first polymers. Future research, guided by the detailed protocols and tools provided here, should continue to integrate these factors, moving from studying them in isolation to exploring their synergistic effects in complex, multi-parameter experiments that more accurately simulate plausible early Earth environments.
Enhancing Replication Fidelity to Maintain Heritable Information
The RNA world hypothesis posits that early life on Earth was based on RNA molecules that served both genetic and catalytic functions, forming the foundation for all subsequent life [23]. Within this framework, the maintenance of heritable information depended fundamentally on the fidelity of RNA replication—the accuracy with which genetic information was copied from one generation to the next. While modern biological systems employ sophisticated protein enzymes to achieve high-fidelity replication, prebiotic systems would have relied on the intrinsic chemical properties of RNA and its precursors [52]. Understanding the mechanisms that enhance replication fidelity in simple RNA-based systems provides crucial insights into the origins of life and has practical implications for modern drug development targeting RNA viruses [86].
The evolutionary success of RNA-based early life depended on achieving a delicate balance: sufficient replication fidelity to maintain functional genetic information, while allowing enough mutation to enable adaptation. RNA viruses today exemplify this balance, operating with error rates of approximately 10⁻⁴ to 10⁻⁶ mutations per round of genome replication [86]. This high mutation rate generates a cloud of closely related virus variants known as a quasispecies, which facilitates rapid adaptation but also positions RNA viruses near an error threshold where further increases in mutation frequency lead to population collapse—a phenomenon termed error catastrophe [86]. This review examines molecular strategies for enhancing replication fidelity, with applications ranging from prebiotic chemistry to antiviral drug development.
Molecular Mechanisms of Replication Fidelity
Template-Directed Synthesis and Base Pairing
In prebiotic RNA replication, heritable information transfer occurred primarily through template-directed synthesis, where existing RNA strands served as templates for complementary strand assembly. The fidelity of this process depended on the thermodynamic stability of Watson-Crick base pairs (G•C and A•U) versus mismatched pairs. Experimental studies suggest that canonical base pairing provides approximately 1-3 kcal/mol additional stability compared to mismatches, creating a natural fidelity mechanism through differential binding affinities [52]. Non-canonical nucleotides, of which approximately 170 have been identified in modern RNA, may have played crucial roles in early replication by modulating base pairing strength and specificity [52].
Ribozyme-Mediated Fidelity Enhancement
The discovery of natural and engineered ribozymes (RNA enzymes) with RNA-dependent RNA polymerase activity provides a plausible mechanism for enhanced replication fidelity in the RNA world. These ribozymes could improve copying accuracy through:
- Geometric selection: Active sites that sterically exclude improperly paired nucleotides
- Transition state stabilization: Preferential stabilization of the chemical transition state for correct versus incorrect nucleotide incorporation
- Proofreading activities: Some ribozymes exhibit nascent strand cleavage activity that removes misincorporated nucleotides
Laboratory evolution experiments have demonstrated RNA polymerase ribozymes capable of synthesizing RNAs longer than themselves, with fidelity sufficient to maintain functional information across generations [23].
Environmental Influences on Fidelity
Prebiotic environmental conditions significantly impacted replication fidelity through multiple factors:
- Temperature moderation: Lower temperatures generally enhance base pairing specificity but slow replication rates
- Divalent cations: Mg²⁺ and other cations facilitate catalysis but can also promote non-enzymatic strand cleavage
- Mineral surfaces: Clay minerals and other surfaces can concentrate nucleotides, align templates, and potentially enhance fidelity through spatial organization [23]
- pH stability: Neutral to slightly acidic pH conditions generally support more accurate replication
The interplay of these factors created environmental niches where fidelity-enhancing conditions prevailed, enabling the emergence and persistence of progressively more complex molecular systems.
Experimental Assessment of Replication Fidelity
In Vitro RNA Replication assays
Protocol: Standard Fidelity Assessment for RNA-Dependent RNA Replication
Template Preparation:
- Synthesize defined RNA templates (200-500 nt) containing specific sequence motifs
- Purify templates by denaturing PAGE or HPLC
- Verify sequence by reverse transcription followed by sequencing
Replication Reaction:
- Assemble 50 μL reactions containing:
- 50 mM HEPES (pH 7.5)
- 150 mM KCl
- 10 mM MgCl₂
- 1 mM each NTP
- 0.5 μM RNA template
- RNA polymerase (ribozyme or protein-based) at appropriate concentration
- Incubate at optimal temperature (typically 20-37°C) for specified time
- Terminate reactions by adding 10 μL 100 mM EDTA
- Assemble 50 μL reactions containing:
Product Analysis:
- Extract RNA with acid phenol:chloroform
- Precipitate with ethanol
- Reverse transcribe using specific primers
- Amplify by PCR
- Clone into sequencing vector
- Sequence 20-50 clones to determine mutation frequency
Fidelity Calculation:
- Calculate mutation rate as (total mutations)/(total nucleotides sequenced)
- Determine sequence-specific error rates by comparing initial and final sequences
Table 1: Key Reagents for RNA Replication Fidelity assays
| Reagent | Function | Considerations |
|---|---|---|
| Defined RNA Templates | Substrate for replication | Should contain reporter regions for fidelity assessment |
| NTPs | Building blocks for RNA synthesis | High-purity grade recommended to prevent incorporation errors |
| Divalent Cations | Cofactors for catalysis | Mg²⁺ most common; Mn²⁺ often increases error rate |
| RNA Polymerase | Catalyzes template-directed synthesis | May be protein-based (modern) or ribozyme (prebiotic models) |
| Reverse Transcriptase | Converts RNA to DNA for analysis | High-fidelity versions recommended to avoid introduction of artifacts |
High-Throughput Fidelity Measurement
Modern approaches employ next-generation sequencing to comprehensively assess replication fidelity:
- Barcode-based Sequencing: Unique molecular identifiers distinguish true mutations from sequencing errors
- Error-Corrected Sequencing: Duplex sequencing methods achieve error rates <10⁻⁷
- Massively Parallel Reporter assays: Assess effects of thousands of mutations simultaneously
These methods provide unprecedented resolution for detecting rare mutations and mapping sequence context effects on fidelity.
Table 2: Quantitative Fidelity Measurements for Representative RNA Replication Systems
| Replication System | Error Rate (mutations/nt) | Key Fidelity Determinants | Experimental Conditions |
|---|---|---|---|
| Qβ Replicase | 1×10⁻⁴ | Template secondary structure, protein fidelity | 37°C, 10 mM Mg²⁺ |
| Poliovirus 3Dpol (wild type) | 3×10⁻⁵ | Active site geometry, conformational selection | 30°C, physiological salt |
| Poliovirus 3Dpol (G64S) | 5×10⁻⁶ | Increased active site selectivity | 30°C, physiological salt [86] |
| RNA Polymerase Ribozyme | 1×10⁻² - 1×10⁻³ | Metal ions, template sequence, selection pressure | Varies by specific ribozyme |
| Non-enzymatic Template Copying | 5×10⁻² - 1×10⁻¹ | Base pairing strength, activated monomers | 0-25°C, prebiotic conditions |
Fidelity-Enhancing Strategies and Their Applications
Genetic and Chemical Approaches
RNA Virus Fidelity Mutants: Site-specific mutations in viral RNA-dependent RNA polymerases can significantly alter replication fidelity. The seminal example is the poliovirus G64S mutation in the RdRp, which decreases error rate approximately 3-fold and creates an attenuated virus with altered tissue tropism [86]. Similar fidelity mutants have been identified in at least 7 RNA virus families, including Picornaviridae, Togaviridae, Flaviviridae, and Coronaviridae [86].
Nucleoside Analogs: Compounds such as ribavirin increase mutation frequency by promoting misincorpororation during RNA replication. When combined with fidelity-modulating mutations, nucleoside analogs can push viral populations past the error threshold into error catastrophe [86]. This approach represents a promising antiviral strategy that directly targets replication fidelity.
Environmental Optimization
Laboratory evolution experiments have identified environmental conditions that enhance replication fidelity:
- Optimized cation mixtures: Specific ratios of Mg²⁺, Mn²⁺, and other cations can improve accuracy
- Molecular crowding agents: Polyethylene glycol and other crowders mimic intracellular conditions and can enhance template binding specificity
- Temperature gradients: Cyclical temperature variations can select for fidelity-enhancing mutations
Directed Evolution of High-Fidelity Systems
Protocol: In Vitro Evolution for Enhanced Fidelity
Starting Population:
- Begin with diverse RNA polymerase ribozyme population (10¹²-10¹⁴ variants)
- Include mutagenic replication conditions to generate diversity
Selection Pressure:
- Implement replication competition under conditions that reward accuracy
- Use templates that require specific sequence maintenance for functional output
- Incorporate counterselection against error-prone variants
Iterative Improvement:
- Cycle through replication and selection phases (20-50 generations)
- Gradually increase fidelity stringency
- Isplicate and characterize improved variants
Characterization:
- Determine error rates for evolved polymerases
- Identify mutations responsible for fidelity enhancement
- Assess trade-offs between fidelity and catalytic efficiency
This approach has yielded ribozymes with significantly improved fidelity, demonstrating the evolutionary accessibility of accuracy enhancement in RNA-based systems.
Visualization of Fidelity Enhancement Concepts
Replication Fidelity Optimization Workflow
RNA Replication Fidelity Experimental Protocol
Research Reagent Solutions for Fidelity Studies
Table 3: Essential Research Reagents for Replication Fidelity Studies
| Reagent/Category | Specific Examples | Function in Fidelity Research |
|---|---|---|
| High-Fidelity Polymerases | poliovirus 3Dpol (G64S mutant), Phi6 RdRp | Engineered viral polymerases with enhanced accuracy for mechanistic studies [86] |
| Ribozyme Polymerases | Class I RNA polymerase ribozyme, R18 polymerase | RNA-based catalysts for prebiotic replication models and origins-of-life research |
| Nucleoside Analogs | Ribavirin, 5-fluorouracil, favipiravir | Chemical mutagens that increase error rates; tools for studying error catastrophe [86] |
| Fidelity Reporters | Luciferase-encoding RNAs, antibiotic resistance genes | Functional assays to quantify phenotypic consequences of mutation rates |
| Template Systems | Defined sequence RNAs, fidelity reporter plasmids | Standardized substrates for comparing fidelity across systems and conditions |
| Mutation Detection Systems | Next-generation sequencing, plaque assay, TLC-based incorporation assays | Tools for quantifying error rates and mutation spectra |
Implications and Future Directions
Enhancing replication fidelity represents a crucial challenge with implications spanning from understanding life's origins to developing novel antiviral strategies. The RNA world hypothesis provides a framework for investigating fundamental principles of information maintenance in simple molecular systems [23]. Meanwhile, research on viral fidelity mutants continues to yield insights with direct therapeutic applications [86].
Future research directions include:
- Engineering ultra-high-fidelity ribozymes to explore the limits of RNA-based information storage
- Developing fidelity-modulating small molecules as broad-spectrum antiviral agents
- Investigating the role of non-canonical nucleotides in primitive replication systems [52]
- Integrating theoretical models with experimental data to predict fidelity thresholds for sustainable replication
These approaches will continue to illuminate the fundamental requirements for maintaining heritable information while providing practical tools for combating RNA-based pathogens through fidelity modulation.
The RNA world hypothesis proposes that early life on Earth was based on RNA molecules that served both genetic and catalytic functions, predating the emergence of DNA and proteins [23] [44]. Within this framework, the ability of RNA to form stable oligomers and higher-order structures was likely essential for the emergence of early evolutionary processes. However, a significant challenge in validating this hypothesis lies in the inherent water instability of RNA and its susceptibility to hydrolysis in prebiotic conditions. Recent research has revealed that stable RNA oligomerization is not merely a historical concept but a functional mechanism observed in modern biological systems, including human pathologies such as cancer metastasis and bacterial stress response [87] [88]. This technical guide synthesizes current research to provide strategies for achieving stable RNA oligomerization, bridging prebiotic chemistry with contemporary biochemical applications. By examining natural instances of RNA-protein co-oligomerization and leveraging advanced experimental technologies, researchers can overcome the water instability problem and unlock new avenues in both origins-of-life research and therapeutic development.
The Fundamental Challenge: Water Instability of RNA
RNA oligomerization in aqueous prebiotic environments faces significant thermodynamic and kinetic barriers. The phosphodiester bonds linking ribonucleotides are inherently susceptible to hydrolytic cleavage in water, especially at elevated temperatures or extreme pH conditions that may have characterized early Earth. This instability contradicts the need for persistent molecular structures capable of storing genetic information and catalyzing reactions.
The search for stable oligomerization mechanisms must account for RNA's structural flexibility and polyanionic nature, which, while enabling functional diversity, also complicate predictable assembly [89]. Beyond prebiotic relevance, this challenge extends to modern therapeutic applications where RNA-targeting small molecules must achieve sufficient binding affinity and specificity amidst RNA's dynamic conformational states [89].
Table: Fundamental Challenges in RNA Oligomerization
| Challenge | Impact on Oligomerization | Potential Solutions |
|---|---|---|
| Hydrolytic instability of phosphodiester bonds | Shortens functional lifespan of oligomers | Seek protective environments; identify stabilizing modifications |
| Structural flexibility and dynamics | Hinders formation of stable intermolecular contacts | Utilize cations or proteins to stabilize specific conformations |
| Polyanionic backbone | Creates charge repulsion between RNA chains | Employ divalent cations (Mg²⁺) or polyamines to screen charge |
| Competition with hydrolysis | Limits thermodynamic driving force for polymerization | Explore dry-wet cycling or mineral surface catalysis |
Natural Paradigms of RNA Oligomerization
tRNA Fragment-Driven Oligomerization in Metastasis
Recent research has revealed that stress-induced transfer RNA fragments (tRFs) can drive functional oligomerization in modern biological systems. A 2022 study demonstrated that a cysteine tRNA fragment (5'-tRFCys) promotes the oligomerization of the RNA-binding protein Nucleolin during breast cancer metastasis [87]. This oligomerization creates a ribonucleoprotein complex that stabilizes specific metabolic mRNAs (Mthfd1l and Pafah1b1), protecting them from exonucleolytic degradation. This mechanism highlights how RNA can act as a structural scaffold facilitating protein oligomerization with significant functional consequences, suggesting possible evolutionary origins where RNA played a similar architectural role.
Nucleotide-Induced Oligomerization in Bacterial Systems
A 2025 study on bacterial transcription termination factor Rho (ρ) revealed that nucleotides can regulate oligomerization states [88]. ADP and the stress-signaling nucleotide (p)ppGpp were found to induce the formation of higher-order ρ oligomers (dodecamers and extended filaments). These oligomerization events inactivate ρ's helicase function by preventing proper ring closure around RNA, representing a regulatory mechanism that responds to cellular stress. This paradigm demonstrates how small molecules can modulate RNA-protein interactions through controlled oligomerization, suggesting similar mechanisms might be exploitable for stabilizing RNA structures.
Figure 1: Nucleotide-induced oligomerization inactivates Rho. Cellular stress signals promote ADP and (p)ppGpp accumulation, which induce Rho filament formation, trapping it in an inactive state [88].
Experimental Strategies for Stable RNA Oligomerization
Structural Characterization Methods
Understanding and engineering stable RNA oligomerization requires sophisticated structural biology approaches. The following table summarizes key methodologies for characterizing RNA structures and oligomerization states:
Table: Experimental Methods for RNA Structure and Oligomerization Analysis
| Method | Application in Oligomerization Studies | Resolution | Throughput |
|---|---|---|---|
| Cryo-EM [89] [88] | Visualization of higher-order oligomers and filaments | Atomic to near-atomic | Medium |
| NMR Spectroscopy [89] [25] | Study of dynamics and small RNA-ligand interactions | Atomic | Low |
| X-ray Crystallography [89] | High-resolution structure of ordered RNA complexes | Atomic | Low |
| Chemical Probing (MaP, DREEM) [89] | RNA folding ensembles and interaction surfaces | Nucleotide | High |
| Crosslinking Assays (BMOE) [88] | Validation of oligomer interfaces in solution | Molecular proximity | Medium |
The Scientist's Toolkit: Essential Research Reagents
Table: Key Reagents for RNA Oligomerization Research
| Reagent/Category | Function in Oligomerization Studies | Specific Examples |
|---|---|---|
| Nucleotide Analogs [88] | Modulate oligomerization states | ADP, (p)ppGpp, ADP•BeF₃ |
| Crosslinkers [88] | Stabilize and detect oligomeric interfaces | Bismaleimidoethane (BMOE) |
| Phosphate Donors [25] | Study prebiotic phosphorylation | Diamidophosphate (DAP) |
| RNA Enrichment Tools [90] | Ispecific RNA species for analysis | Chaplet chromatography, DNA nanoswitches |
| Stabilizing Cations | Counteract charge repulsion in RNA backbone | Mg²⁺, polyamines |
Detailed Experimental Protocol: Analyzing tRNA Fragment-Induced Oligomerization
Based on the study of pro-metastatic tRNA fragments [87], the following protocol outlines key methodology for investigating RNA-driven oligomerization:
Objective: To determine whether a specific tRNA fragment (5'-tRFCys) drives Nucleolin oligomerization and stabilizes bound mRNAs.
Procedure:
Small RNA Profiling: Extract total RNA from cancer cell models (e.g., metastatic breast cancer cells) using guanidinium thiocyanate-based methods. Isolate small RNA fractions (<200 nt) and sequence using high-throughput platforms to identify differentially expressed tRFs, particularly 5'-tRFCys.
Functional Validation of 5'-tRFCys:
- Knock down 5'-tRFCys using antisense oligonucleotides (ASOs) or CRISPR-based approaches.
- Assess functional outcomes through metastasis assays (e.g., transwell migration, lung colonization in animal models).
- Evaluate cancer cell survival under stress conditions using viability assays.
Identification of RNA-Binding Protein Partners:
- Perform RNA pull-down assays using biotinylated 5'-tRFCys and cell lysates.
- Identify bound proteins through mass spectrometry, with Nucleolin as a candidate.
- Validate direct binding using surface plasmon resonance (SPR) or electrophoretic mobility shift assays (EMSAs).
Oligomerization Analysis:
- Incubate purified Nucleolin with 5'-tRFCys in physiological buffer.
- Analyze oligomer formation through:
- Size-exclusion chromatography with multi-angle light scattering (SEC-MALS)
- Crosslinking with BMOE followed by SDS-PAGE [88]
- Pelleting assays (100,000 × g) to sediment large oligomers
- Native PAGE to resolve different oligomeric states
mRNA Stabilization Assessment:
- Identify Nucleolin-bound metabolic mRNAs (e.g., Mthfd1l, Pafah1b1) through RNA immunoprecipitation sequencing (RIP-seq).
- Measure mRNA half-lives after transcription inhibition with actinomycin D.
- Quantify exonuclease resistance of target mRNAs in the ribonucleoprotein complex.
Figure 2: Experimental workflow for studying RNA-driven oligomerization. The process begins with RNA profiling and progresses through functional validation to mechanistic analysis of oligomerization and its functional consequences [87].
Computational Approaches for Rational Design
Advancements in computational methods have dramatically improved our ability to predict and design stable RNA oligomers:
RNA Structure Prediction: Machine learning algorithms, particularly deep learning approaches, can now predict RNA secondary and tertiary structures with remarkable accuracy by integrating sequence information, chemical probing data, and evolutionary conservation [89]. These tools help identify potential oligomerization interfaces by modeling RNA's conformational landscape.
Molecular Dynamics Simulations: Physics-based modeling can simulate RNA folding and oligomerization processes at atomic resolution, providing insights into stability under various environmental conditions [89]. Specialized force fields account for RNA's unique electrostatic properties and hydration effects.
Rational Ligand Design: Computational screening of small molecules that stabilize RNA oligomers can identify chemical scaffolds that promote specific quaternary structures [89]. Fragment-based drug discovery approaches are particularly promising for exploring the chemical space of RNA binders.
Implications for the RNA World Hypothesis
The natural paradigms of RNA oligomerization observed in contemporary biology provide valuable insights for prebiotic chemistry research. The discovery that tRNA fragments can drive functional protein oligomerization [87] suggests a possible mechanism by which early RNA molecules might have coordinated the assembly of primitive molecular machines before the emergence of sophisticated protein-based enzymes.
Recent research on ribose selection provides clues about how nature might have chosen specific molecular components long before enzymes existed [25]. Studies show that ribose binds more readily and selectively to phosphate compared to other similar sugars, forming structures ideal for RNA formation [25]. This inherent chemical preference could have facilitated the emergence of the first stable RNA oligomers.
The observation that nucleotides can regulate oligomerization states [88] suggests a primitive regulatory mechanism that might have operated in early RNA-world organisms. Small molecules could have modulated RNA function by controlling its assembly into higher-order structures, providing a basic system of metabolic control before the evolution of protein-based allosteric regulation.
Future Directions and Applications
Prebiotic Chemistry Research: Future work should focus on reconstructing plausible prebiotic scenarios that enable stable RNA oligomerization. This includes investigating mineral surface catalysis, dry-wet cycling environments, and potential early cellular compartments (protocells) that might have protected emerging RNA oligomers from hydrolysis [25].
Therapeutic Development: Understanding natural RNA oligomerization mechanisms opens new avenues for drug discovery. Targeted RNA degraders and small molecules modulating RNA-protein interactions represent promising therapeutic strategies for diseases ranging from cancer to neurological disorders [89].
Technology Development: The emerging Human RNome Project aims to comprehensively map RNA modifications and structures across cell types [90]. This initiative will provide crucial data for understanding the structural principles governing RNA oligomerization and developing new strategies to control it.
The convergence of structural biology, computational modeling, and chemical biology continues to provide new strategies for overcoming the water instability problem of RNA oligomerization. By learning from natural paradigms and developing innovative experimental approaches, researchers are building a more comprehensive understanding of how RNA could have served as the cornerstone of early life while opening new possibilities for RNA-based therapeutics.
The transition from a prebiotic chemical environment to the first biological systems required pathways for the specific synthesis of peptides. Within the framework of the RNA world hypothesis, a major challenge is explaining how selective peptide bond formation occurred prior to the evolution of the complex ribosomal machinery. This whitepaper details prebiotically plausible chemical mechanisms that achieve highly selective α-amino acid coupling over other competing amines. We examine the critical role of pH, specific activating agents, and the formation of cyclic intermediates in enforcing selectivity. Furthermore, we explore the emergence of an RNA-peptide world, where the intrinsic chemistry of RNA nucleosides provided a scaffold for the earliest peptide synthesis. The experimental data and protocols herein provide a foundational guide for researchers investigating the chemical origins of life and for drug development professionals seeking to exploit non-enzymatic coupling strategies.
The RNA world hypothesis posits that an early stage in the evolution of life was dominated by self-replicating RNA molecules, which stored genetic information and catalyzed chemical reactions [81] [38]. A central paradox in this hypothesis is how this world evolved into one where proteins became the dominant catalysts, given that protein synthesis itself is catalyzed by the RNA-based ribosome. This presents a "chicken-and-egg" conundrum: which came first, the ribosome capable of making proteins, or the proteins required to build the ribosome? [24].
A critical step in resolving this paradox is identifying prebiotically plausible mechanisms for selective peptide bond formation. The early Earth likely contained a complex mixture of molecules, including various α-amino acids, β-amino acids, diamines, and other nucleophiles [91]. For functional peptides to emerge, condensation reactions needed to favor α-amino acids, the building blocks of modern proteins. However, peptide bond formation faces significant thermodynamic barriers in water, and achieving selectivity without modern enzymatic control is chemically challenging [92]. This technical guide explores the specific chemical conditions—including pH, activating agents, and intermediate structures—that enable such selectivity, providing a plausible pathway for the emergence of the first peptides within an RNA-peptide world [93].
Core Chemical Mechanisms for Selective Coupling
pH-Dependent Selection via Mixed Anhydride Intermediates
A seminal study demonstrated that pH control is a powerful tool for achieving selective peptide elongation. Using methyl isonitrile as a prebiotically plausible activating agent, researchers found that the reactivity of different amine nucleophiles varies dramatically with pH [91].
At pH 5, the reaction shows limited selectivity; glycine, β-alanine, glycylglycine, and methylamine all form amide bonds with a model peptide (N-acetyl-L-alanine) in comparable yields. However, at pH 3, a strong preference for α-amino acids emerges. In a competitive experiment containing multiple amines, glycine was incorporated into peptides with 55% yield, while the reaction of β-alanine was fully suppressed [91].
The proposed mechanism for this selectivity involves a switch from a direct intermolecular pathway to an intramolecular O→N acyl transfer [91]. This indirect pathway is unique to α-amino acids and becomes dominant under acidic conditions:
- The carboxylate of the N-acetylated peptide chain is activated by methyl isonitrile, forming an imidoyl acetyl alanine intermediate.
- Instead of a direct attack by an external amine (which is protonated and unreactive at low pH), the carboxylate group of the incoming α-amino acid attacks the activated intermediate, forming a mixed carboxylic-carbonic anhydride intermediate.
- This mixed anhydride undergoes a facile intramolecular rearrangement through a 5-membered ring transition state, transferring the acyl group to the α-amino group of the same molecule and forming the peptide bond.
This mechanism leverages the bifunctional nature of α-amino acids and the lower entropic penalty of intramolecular reactions to achieve selectivity under conditions where most competing amines are protonated and inert [91]. The following diagram illustrates this selective pathway.
RNA-Templated Peptide Synthesis Using Non-Canonical Nucleosides
An alternative and highly innovative pathway to selectivity involves the direct growth of peptides on RNA scaffolds, a scenario central to the RNA-peptide world hypothesis. This mechanism utilizes non-canonical nucleosides, considered molecular fossils from the RNA world, which are found in modern transfer and ribosomal RNAs [93].
These nucleosides, such as N6-methyl-N6-threonylcarbamoyladenosine (m6t6A) and 5-methylaminomethyluridine (mnm5U), inherently contain amino acid residues or modified side chains that can participate in chemistry. When positioned close to each other on complementary RNA strands, they can directly mediate peptide bond formation without the need for a modern ribosome [93].
The stepwise mechanism for this process is as follows:
- Hybridization & Activation: An RNA "donor" strand containing an m6aa6A nucleotide (where 'aa' is an amino acid) is hybridized to an RNA "acceptor" strand containing an (m)nm5U nucleotide at its 3' end. The carboxylic acid of the donor's amino acid is activated.
- Peptide Bond Formation: The nucleophilic amine from the acceptor strand's amino acid moiety attacks the activated carbonyl on the donor strand, forming a new peptide bond and creating a hairpin-type RNA-peptide chimera.
- Urea Cleavage & Strand Displacement: The urea linkage connecting the peptide to the m6A nucleobase is cleaved, releasing the peptide onto the acceptor strand. A new donor strand can then hybridize, allowing for iterative peptide elongation.
This process demonstrates inherent amino acid selectivity, as the coupling rate (kapp) varies significantly with the identity of the amino acid (e.g., phenylalanine couples much faster than glycine), likely due to differential pre-organization on the RNA scaffold [93]. The process of peptide growth on an RNA scaffold is detailed below.
Quantitative Data and Experimental Comparisons
The following tables summarize key quantitative findings from the primary research discussed, providing a clear comparison of the factors governing selectivity.
Table 1: Amine Nucleophile Incorporation Yield at Different pH Levels with Methyl Isonitrile Activation [91]
| Amine Nucleophile | pKaH of Amine | Yield at pH 5 | Yield at pH 4 | Yield at pH 3 |
|---|---|---|---|---|
| Glycine (3a) | 9.6 | 62% | 58% | 46% |
| β-Alanine (3b) | 10.2 | Comparable to 3a | 47% | 5% |
| Glycylglycine (3c) | 8.1 | Comparable to 3a | ~20% | 0% |
| Glycine nitrile (3e) | 5.2 | Quantitative | Quantitative | 30% |
| Methylamine (3f) | 10.6 | Comparable to 3a | 0% | 0% |
Table 2: Coupling Efficiency of Different Amino Acids on an RNA Scaffold [93]
| Amino Acid (in Donor Strand) | Apparent Rate Constant (kapp, h⁻¹) | Relative Coupling Rate |
|---|---|---|
| Phenylalanine (F) | > 1.0 | Highest |
| Leucine (L) | ~0.4 | High |
| Threonine (T) | ~0.4 | High |
| Methionine (M) | ~0.4 | High |
| Glycine (G) | ~0.1 | Low |
Detailed Experimental Protocols
Protocol 1: pH-Dependent Selective Coupling with Methyl Isonitrile
This protocol is adapted from the work demonstrating selective α-amino acid coupling under acidic aqueous conditions [91].
Research Reagent Solutions
| Reagent | Function & Note |
|---|---|
| Methyl isonitrile (1) | Prebiotic activating agent. Handle in fume hood. |
| N-acetyl-L-alanine (2) | Model peptide precursor. |
| α-Amino acids (e.g., Gly, Arg, Ser, Val, Pro) | Primary amine nucleophiles for coupling. |
| Phosphate or Formate Buffers | For maintaining precise pH (3, 4, and 5). |
| D₂O | Solvent for reaction monitoring via ¹H-NMR. |
Methodology:
- Reaction Setup: Prepare an aqueous solution of N-acetyl-L-alanine (2, 100 mM) and methyl isonitrile (1, ~130 mM) in the appropriate buffer.
- Nucleophile Addition: Add the amine nucleophile (e.g., glycine, 50 mM) to the mixture. For competition experiments, prepare a mixture of multiple amines (e.g., 25 mM each of glycine, β-alanine, glycylglycine, etc.).
- Incubation: Incubate the reaction mixture at 23°C (room temperature) and monitor by ¹H-NMR spectroscopy until the methyl isonitrile is fully consumed. The reaction time is highly pH-dependent:
- pH 3: ~6 hours
- pH 4: ~48 hours
- pH 5: ~15 days
- Analysis: Identify and quantify the formation of elongated peptide products (e.g., Ac-Ala-Gly) by integrating characteristic peaks in the ¹H-NMR spectrum. Yields are calculated based on the initial concentration of the amine nucleophile.
Protocol 2: RNA-Templated Peptide Synthesis via Non-Canonical Nucleosides
This protocol is based on the "palaeochemistry" approach that forms peptide bonds directly on RNA scaffolds [93].
Research Reagent Solutions
| Reagent | Function & Note |
|---|---|
| RNA Donor Strands (e.g., 1a-1j) | Contain m6aa6A nucleotides at 5' end. |
| RNA Acceptor Strands (e.g., 2a-2c) | Contain (m)nm5U nucleotides at 3' terminus. |
| EDC / Sulfo-NHS or DMTMM·Cl | Carboxylic acid activating agents. |
| Methyl isonitrile | Alternative prebiotic activator (pH 6). |
| DTT (Dithiothreitol) | For thiol-activation chemistry with nitrile derivatives. |
| 2'-OMe Nucleotides | Used to enhance RNA stability under harsh conditions. |
Methodology:
- RNA Synthesis and Purification: Synthesize the donor and acceptor RNA strands (e.g., 5- to 9-mers) using solid-phase synthesis, incorporating the specified non-canonical nucleosides (m6aa6A, nm5U). Purify the strands via HPLC.
- Hybridization: Mix the donor and acceptor RNA strands in an equimolar ratio in a buffer (e.g., 50 mM MES, pH 6.0). Heat the mixture to 65°C for 5 minutes and allow it to cool slowly to 25°C to facilitate hybridization.
- Coupling Reaction: Activate the carboxylic acid of the donor strand. Two primary methods are used:
- Chemical Activation: Add an activating agent like EDC (with Sulfo-NHS) or DMTMM·Cl to the hybridized RNA solution.
- Prebiotic Thiol Activation: For donor strands with amino acid nitriles (e.g., 1j), use DTT (50 mM) at pH 8.
- Incubation and Monitoring: Incubate the reaction at 25°C. Monitor the formation of the hairpin-type RNA-peptide chimera and subsequent products using analytical HPLC or LC-MS over several hours.
- Urea Cleavage (Optional): To release the peptide onto the acceptor strand for further elongation, incubate the chimera at elevated temperature (90°C) and mildly acidic pH (4-6) for several hours to cleave the urea linkage.
- Fragment Coupling (For Longer Peptides): To form longer peptides, use a pre-formed peptide-coupled adenosine nucleoside ((m6)peptide6A) incorporated into a new donor strand. Hybridize this with the peptide-decorated acceptor strand from a previous cycle and repeat the activation and coupling steps.
The Scientist's Toolkit: Essential Research Reagents
The following table catalogues key reagents and their functions in prebiotic peptide coupling experiments, serving as a quick reference for researchers designing related studies.
Table 3: Key Reagent Solutions for Prebiotic Coupling Experiments
| Reagent / Chemical | Function in Experimental Context | Prebiotic Plausibility & Notes |
|---|---|---|
| Methyl Isonitrile | Activates carboxyl groups, enabling peptide bond formation in water [91]. | Considered a plausible prebiotic molecule available on early Earth [91]. |
| Non-Canonical Nucleosides (e.g., m6t6A, mnm5U) | Act as built-in amino acid carriers and catalysts for peptide synthesis on RNA scaffolds [93]. | Considered "molecular fossils"; found in modern tRNA/rRNA, indicating an ancient origin. |
| α-Hydroxy Acids (e.g., l-lactic acid) | Co-polymerize with α-amino acids to form depsipeptides (polyester-amide hybrids) [92]. | Found in meteorites and laboratory prebiotic synthesis experiments; proposed as peptide precursors. |
| Amino Acid Analogues (e.g., α-aminonitriles) | Serve as activated monomers that facilitate peptide coupling under milder conditions [91] [92]. | Proposed intermediates in prebiotic amino acid synthesis pathways like the Strecker reaction. |
| Wet-Dry Cycles | A physical process that concentrates reactants, drives condensation, and promotes chemical evolution [92] [93]. | Highly plausible geological scenario on early Earth (e.g., at tidal pools or hot springs). |
The experimental evidence confirms that specific peptide coupling without modern enzymes is achievable through well-defined chemical principles. The strategies outlined—pH-dependent selectivity and RNA-templated synthesis—provide robust, prebiotically plausible pathways that align with and enrich the RNA world hypothesis. They demonstrate how the unique chemical properties of α-amino acids and RNA nucleosides could have been leveraged to kickstart the production of functional peptides.
Future research should focus on exploring a wider range of prebiotic activating agents and environmental conditions, such as temperature and ionic strength. A key challenge is demonstrating the synthesis of longer, functionally catalytic peptides using these methods. Furthermore, the integration of these peptide-forming reactions with systems capable of RNA replication will be crucial for building a comprehensive model of a evolving RNA-peptide world. For drug development, these non-enzymatic, chemoselective strategies offer inspiration for novel bioconjugation techniques and the synthesis of complex biomimetic polymers under green and sustainable conditions.
The origin of life on Earth presents a fundamental chemical paradox: the biomolecules essential for life, particularly nucleotides and RNA, require specific environmental conditions for stabilization and interaction, yet the primordial oceans were vast and dilute [24]. Within the framework of the RNA World Hypothesis, which posits that RNA served as both the primary genetic material and catalytic molecule in early evolution, this concentration challenge becomes particularly critical [8] [24]. The hypothesis suggests that RNA was a key player during a stage of evolution before the emergence of the contemporary DNA/RNA/protein world [24]. However, the prebiotic synthesis of RNA faces significant objections, including the inherent instability of RNA and the complexity of its constituent nucleotides, which are difficult to form under plausible early Earth conditions [8] [94] [24]. This technical guide explores the experimental approaches and environmental simulations designed to overcome the dilution problem, providing a detailed analysis for researchers investigating the chemical origins of life.
The core of the challenge lies in the properties of RNA itself. RNA is a complex molecule, susceptible to hydrolysis, especially at neutral to alkaline pH, and its catalytic activities are often dependent on specific folding and the presence of stabilizers like certain ions [24]. For instance, while Mg²⁺ is crucial for stabilizing RNA structure, high concentrations can also catalyze its degradation, creating a delicate balancing act for researchers designing experiments [24]. Furthermore, the prebiotic chemistry processes that could lead to the formation of RNA's building blocks—a focus of fields aiming to understand the formation of life's essential molecules like amino acids and nucleotides—would have been inefficient in a vast, dilute ocean [94]. Consequently, the scientific community has investigated various geological settings that could have provided a concentrated environment, acting as a crucible for the emergence of the first self-replicating systems.
Environmental Scenarios for Concentration
Experimental simulations have focused on several key environments that could have provided the necessary conditions for concentrating prebiotic reagents and facilitating RNA formation and stability. The quantitative parameters associated with these environments are summarized in Table 1 below.
Table 1: Quantitative Parameters for Simulated Prebiotic Concentration Environments
| Environmental Scenario | Key Concentration Mechanism | Typical Experimental pH Range | Temperature Range | Key Stabilizing Factors | Primary Experimental Challenges |
|---|---|---|---|---|---|
| Ice Eutectic Phases | Freeze-concentration of solutes in liquid veins within ice | Varies, often near neutral | -7°C to -8°C (for maximal reported ribozyme activity) [24] | Increased molecular crowding, lowered water activity [24] | Increased intermolecular base-pairing can reduce catalytic activity [24] |
| Hydrothermal Vents | Evaporative concentration, thermal gradients, mineral catalysis | Acidic (pH 4–5) for RNA stability [24]; Alkaline (pH 9–11) for some metabolism-first models [24] | 40°C to 70°C (for some DNA synthesis pathways) [21]; higher for vent fluids | Mineral surfaces (e.g., clay), protonated nucleotides at low pH [24] | High temperatures can accelerate RNA degradation; requires precise thermal control |
| Freshwater Pools & Lakes | Evaporative concentration, wet-dry cycles, mineral binding | Acidic (pH 4–5) for RNA backbone stability [24] | Ambient to elevated (e.g., 40-70°C for lab simulations) | Clay minerals, lipids for compartmentalization | Simulating realistic seasonal and diurnal cycles in the lab |
As detailed in Table 1, each proposed environment offers a distinct pathway to concentration. The ice eutectic phase model leverages the natural exclusion of solutes from crystal structures during freezing, leading to a significant increase in the concentration of molecules in the remaining liquid pockets. Research has demonstrated that some ribozymes exhibit maximal activity at temperatures of -7°C to -8°C, attributed to this concentration effect and reduced water activity [24]. Conversely, hydrothermal vent scenarios utilize thermal and pH gradients to drive chemical reactions. A significant body of work suggests that an acidic pH (4-5) is beneficial for RNA stability, as it protects the phosphodiester backbone from base-catalyzed hydrolysis and can reduce the dependency on Mg²⁺, which is both a stabilizer and a potential catalyst of degradation [24]. Finally, freshwater pools and tidal basins are simulated through repeated wet-dry cycles, which promote condensation reactions and polymerization by removing water and concentrating reactants on mineral surfaces like clay.
Experimental Protocols for Simulating Concentration
This section provides detailed methodologies for key experiments that simulate concentration mechanisms in a laboratory setting, providing a reproducible framework for investigating prebiotic RNA chemistry.
Protocol A: Ice Eutectic Phase Simulation for Ribozyme Studies
This protocol is designed to test RNA activity and stability under icy conditions, relevant to both early Earth and icy moons like Europa [24].
- Solution Preparation: Prepare a buffered solution containing the RNA molecule of interest (e.g., a self-splicing ribozyme), along with necessary ions (e.g., Mg²⁺ at carefully titrated concentrations). The buffer should maintain a pH between 5 and 6 to minimize RNA hydrolysis [24].
- Incubation and Freezing: Aliquot the solution into sterile vials. Incubate the vials in a precision temperature bath set to a target sub-zero temperature, typically within the range of -7°C to -8°C, where maximal ribozyme activity has been observed [24].
- Activity Assay: After a set period, rapidly thaw the samples on ice. Measure ribozyme activity using a standardized biochemical assay. This often involves providing a fluorescently quenched substrate; cleavage by the ribozyme results in a fluorescent signal quantifiable by a plate reader.
- Stability Analysis: Use techniques like denaturing gel electrophoresis (e.g., urea-PAGE) to assess RNA integrity post-incubation, looking for signs of degradation.
Protocol B: Acidic Hydrothermal Conditions for RNA Stabilization
This protocol investigates RNA polymerization and stability in simulated acidic hydrothermal environments, which may have been common on the early Earth due to volcanic outgassing [24].
- Reactor Setup: Utilize a chemical flow reactor system that can maintain a stable temperature gradient (e.g., 40°C to 90°C) and a constant acidic pH, ideally between 4 and 5 [24].
- Nucleotide Introduction: Introduce a solution of activated nucleotides (e.g., phosphorimidazolides of adenosine) into the reactor. The solution may include mineral catalysts such as montmorillonite clay, known to facilitate RNA oligomerization.
- Template-Directed Polymerization: Include a short RNA primer and a complementary RNA template to study non-enzymatic replication. The flow system allows for continuous replenishment of reagents.
- Product Analysis: Collect effluent from the reactor and analyze the products using HPLC-MS (High-Performance Liquid Chromatography-Mass Spectrometry). This identifies the length and sequence of the synthesized RNA oligomers.
Protocol C: Wet-Dry Cycling in Freshwater Simulants
This protocol mimics the concentration effects of evaporating tidal pools or freshwater lakes, a widely studied model for polymerization.
- Mineral Substrate Preparation: Use a flat substrate such as a glass slide or a bed of powdered clay minerals (e.g., kaolinite or montmorillonite).
- Reagent Application: Apply an aqueous solution containing nucleotides, amino acids, or other prebiotic building blocks to the substrate.
- Cycling Regime: Place the substrate in an environmental chamber that cycles between hydration (high humidity or direct application of water) and dehydration (elevated temperature, e.g., 40-70°C, and low humidity). Each cycle can last 12-24 hours.
- Product Characterization: After a set number of cycles (e.g., 10-50), wash the substrate to collect synthesized products. Analyze polymers using gel electrophoresis for nucleic acids or chromatography for peptides.
The logical relationship between the experimental challenge of a dilute prebiotic soup and the simulated environments that offer solutions is visualized in the following workflow.
The Scientist's Toolkit: Research Reagent Solutions
Successfully simulating prebiotic concentration environments requires a specific set of chemical and analytical tools. The table below details essential reagents and materials, their functions, and relevant experimental considerations.
Table 2: Key Research Reagents and Materials for Prebiotic Concentration Experiments
| Reagent/Material | Core Function | Application Example | Stability & Handling Notes |
|---|---|---|---|
| Ribozymes (e.g., R3C Ligase) | Catalytic RNA model; demonstrates RNA's dual role as gene and enzyme [12]. | Study of allosteric regulation and stability in different concentrated environments (e.g., ice, acidic pH) [12]. | RNase-free conditions are critical; stable at acidic pH but labile at high pH and temperature [24]. |
| Activated Nucleotides | Monomers for non-enzymatic RNA polymerization; contain leaving group for reaction. | Template-directed RNA copying in wet-dry cycles or on mineral surfaces [24]. | Often moisture-sensitive; store desiccated at -20°C; use imidazolide or other prebiotically plausible activators. |
| Clay Minerals (Montmorillonite) | Natural catalyst; provides surface for adsorption and concentration of organics. | Polymerization of amino acids or nucleotides in simulated freshwater pools [94]. | Variable cation exchange capacity; should be characterized (e.g., XRD) before use. |
| Magnesium Ions (Mg²⁺) | Cofactor for RNA folding and ribozyme function; stabilizes tertiary structure. | Essential component in ribozyme activity buffers for ice eutectic and other experiments [24]. | Concentration must be optimized; high levels catalyze RNA strand cleavage [24]. |
| Deoxyribonucleoside Precursors | Prebiotic precursors for DNA subunits (e.g., acetaldehyde, glyceraldehyde). | Studying parallel emergence of DNA and RNA in mildly alkaline hydrothermal conditions [21]. | Reactions often require controlled temperature (40-70°C) and pH [21]. |
Advanced Techniques and Recent Experimental Findings
Cutting-edge research in prebiotic chemistry continues to refine our understanding of concentration mechanisms and their implications for the RNA World. Advanced experimental setups now allow for more sophisticated simulations and analyses.
Allosteric Regulation in Engineered Ribozymes
Recent work has demonstrated that ribozymes can be engineered to be allosterically regulated by key biomolecules, a property that may have deep evolutionary roots. A 2024 study engineered the R3C ligase ribozyme by fusing it with short RNA sequences that bind ATP or L-histidine [12]. The researchers observed that the ligase activity became dependent on the concentration of these effector molecules. Higher concentrations of ATP or L-histidine led to increased ligase activity and structural stabilization, as indicated by an increase in melting temperature (Tm) [12]. This finding suggests that early RNA systems could have developed complex regulatory networks in concentrated microenvironments, where such effectors were available, potentially bridging the RNA World to the modern "DNA/Protein World."
Parallel Reaction Monitoring in Complex Networks
To unravel the complex reaction networks that constitute prebiotic chemistry, advanced analytical techniques are required. One prominent method is the use of parallel reactors with continuous reaction progress analysis [21]. This sophisticated approach involves:
- Hardware: An array of small-volume reactors housed in a system that allows for precise control of temperature, pressure, and pH.
- Process: The continuous or semi-continuous monitoring of complex reaction mixtures as they evolve over time.
- Analysis: Integration of this system with high-throughput analytical techniques, such as liquid chromatography-mass spectrometry (LC-MS), to track the formation and consumption of hundreds of chemical species simultaneously.
- Objective: The primary goal is to identify catalytic components, selective interactions, and missing reaction pathways within the network that lead to the synthesis of key biopolymers like RNA [21]. This methodology is particularly powerful for identifying cooperative effects that drive the system toward higher complexity.
The RNA-Peptide World and Co-Evolution
The traditional "RNA-first" view is being challenged and complemented by the "RNA-Peptide World" hypothesis, which proposes that RNA and peptides co-evolved from the very beginning. A 2022 study presented a prebiotically plausible scenario where RNA molecules with non-canonical nucleotides directly templated the growth of peptides [95]. In this model, the stronger chemical bonds in these RNA-peptide chimeras provided stability in water. Crucially, the resulting peptides, in turn, stabilized the cooperating RNA molecules, creating a positive feedback loop [95]. This synergy, likely occurring in a protected, concentrated setting, would have allowed both classes of molecules to evolve towards greater complexity, eventually leading to the separation of functions seen in modern biology. The experimental workflow for studying such chimera formation is visualized below.
The challenge of concentration is not merely a technical obstacle in prebiotic simulations but a central conceptual problem in understanding the origin of life. Experimental evidence strongly indicates that the dilute ocean was an improbable cradle for life's genesis; instead, localized microenvironments like icy eutectic phases, acidic hydrothermal vents, and freshwater pools provided the necessary conditions to concentrate reagents, promote polymerization, and stabilize fragile biomolecules like RNA. While the RNA World Hypothesis has been aptly described as "the worst theory of the early evolution of life (except for all the others)" due to its unresolved challenges, research into concentration mechanisms provides a critical pathway for validating and refining it [8] [24]. The emerging paradigm of an RNA-Peptide World, where both molecules co-evolved in a synergistic relationship within concentrated niches, offers a compelling and experimentally tractable framework for future research [95]. For scientists in biophysics and drug development, these natural concentration strategies offer inspiration for improving the efficiency of oligonucleotide synthesis and the design of functional nucleic acids, bridging the gap between the origins of life and modern biotechnological applications.
Validating the Hypothesis: Ancient Biosignatures and Comparative Analysis
The quest to understand the origins of life presents one of science's most profound challenges, central to which is the RNA world hypothesis. This concept posits that early life on Earth was based on RNA molecules capable of both storing genetic information and catalyzing chemical reactions, predating the DNA-protein world we know today [24] [23]. A critical, yet elusive, piece of evidence for this theory lies in the detection of chemical fossils—molecular biosignatures preserved in ancient rocks that can provide a direct record of primordial biological activity. For decades, the study of such ancient biosignatures has been constrained by the degradation of these molecular structures over geological timescales and the limitations of analytical techniques to identify faint, residual traces.
Recent advances in machine learning (ML) are now revolutionizing this field. By applying sophisticated pattern recognition to complex chemical data, researchers can now detect the subtle "chemical echoes" of ancient life, even when the original biomolecules are no longer intact [96]. This technical guide explores the integration of machine learning with analytical chemistry to detect ancient biosignatures, framing these advancements within the broader context of prebiotic chemistry and the search for evidence supporting the RNA world hypothesis.
Chemical Fossils and the RNA World
The Nature of Chemical Fossils
Chemical fossils, or molecular biosignatures, are the diagenetic remnants of biological lipids, pigments, and other structural components. Unlike body fossils, which preserve the shape of an organism, chemical fossils preserve the molecular and isotopic evidence of past life. In the context of the RNA world, this evidence could include the molecular fossils of noncanonical RNA nucleosides.
A foundational 2018 study demonstrated that methylated and carbamoylated RNA nucleosides, which are still present in modern RNA, can be generated through prebiotic chemistry involving isocyanates and sodium nitrite. This provides a plausible scenario for the chemical origin of certain noncanonical bases, suggesting they are molecular fossils of an early Earth [97]. The detection of such compounds in ancient geological samples would provide powerful, direct support for the RNA world hypothesis.
Challenges in Traditional Detection Methods
The primary obstacle in studying these biosignatures is their degradation over time. Geological processes subject rocks to immense heat and pressure, destroying most original biomolecules [96]. While larger biomarker molecules degrade, their smaller, more stable molecular fragments persist. Traditional analytical methods, like gas chromatography-mass spectrometry (GC-MS), have historically struggled to reliably connect these fragment patterns to a biological origin, especially in samples older than 1.7 billion years [98].
Machine Learning-Driven Detection Framework
Core Methodology: Pyrolysis GC/MS and Molecular Fragment Analysis
The breakthrough in detection leverages a core analytical technique—pyrolysis gas chromatography/mass spectrometry (Py-GC/MS)—enhanced by machine learning.
- Pyrolysis GC/MS Workflow: This method involves heating rock samples to high temperatures in an inert atmosphere, causing them to break down into smaller, volatile molecular fragments. These fragments are then separated by gas chromatography and identified by mass spectrometry, producing a complex dataset that represents the sample's molecular fingerprint [98].
- The Machine Learning Advantage: Instead of searching for a single, specific biomarker molecule (which may have degraded beyond detection), the ML model is trained to recognize the overall pattern of fragments characteristic of biological origins. As researcher Michael Wong describes, "It's basically analogous to facial recognition, except instead of facial features, it’s looking at these chemical datasets" [98].
The following diagram illustrates the complete experimental workflow, from sample preparation to biological classification.
Model Training and Performance
The efficacy of this approach hinges on robust training data. A landmark 2025 study by Hazen and colleagues built a model using a diverse reference set of 406 samples, including modern plants, billion-year-old fossils, and non-biological materials like meteorites [98] [96].
Table 1: Machine Learning Model Performance on Reference Data
| Sample Type | Model Classification Accuracy | Key Findings/Challenges |
|---|---|---|
| Overall Biological vs. Non-Biological | >90% [98] [96] | Model distinguishes with high reliability. |
| General Ancient Rocks | Successful detection in ~3.3 billion-year-old samples [96] | Pushes back chemical evidence of life by ~1.6 billion years. |
| Photosynthetic Organisms | Detected in ~2.5 billion-year-old samples [98] [96] | Extends record of photosynthesis by ~800 million years. |
| Closely Related Specimens | ~79% [98] | Challenging to distinguish, e.g., photosynthetic vs. non-photosynthetic plants. |
This data demonstrates the model's high accuracy while also highlighting areas for improvement, primarily through the expansion of training datasets to include more diverse and complex samples.
Essential Research Reagents and Materials
Implementing this detection framework requires specific reagents and analytical tools. The following table details key components used in the featured experiments.
Table 2: Research Reagent Solutions for ML-Driven Biosignature Detection
| Item Name | Function/Description | Role in the Experimental Protocol |
|---|---|---|
| Pyrolysis GC/MS System | An instrument that thermally decomposes a sample and separates/identifies the resulting fragments. | Core analytical platform for generating molecular fragment data from solid rock samples [98]. |
| Diverse Reference Sample Set | A curated collection of modern biological, ancient fossil, and non-biological (e.g., meteorite) materials. | Used to train the machine learning model to recognize diagnostic chemical patterns [98] [96]. |
| Machine Learning Model (Algorithm) | A computational model (e.g., based on chemical pattern recognition). | Analyzes complex pyrolysis GC/MS data to classify sample origin [98]. |
| Ancient Rock Thin Sections | Slices of rock mounted on slides for analysis. | The primary source material for analysis; can be sourced from museum collections with minimal destruction [99]. |
| Infrared Spectrometer | An instrument that measures the interaction of infrared light with a sample's molecular bonds. | An alternative/complementary technique to Py-GC/MS that provides molecular data with minimal sample preparation [99]. |
Molecular Representation Learning for Advanced Analysis
Beyond identifying the presence of life, new computational methods are being developed to understand the intricate structures of ancient biomolecules. Molecular Representation Learning (MRL) is a frontier field in machine learning that translates molecular structures into numerical representations that computers can process for predictive tasks [100].
A key challenge is moving beyond simple molecular representations like SMILES (Simplified Molecular Input Line Entry System), which struggle with complex chemical entities like the Markush structures common in patents and potentially relevant to prebiotic chemistry [101]. Innovative frameworks like Knowledge-aware Contrastive Heterogeneous Molecular Graph Learning (KCHML) address this by representing a molecule as a heterogeneous graph, integrating multiple views:
- Molecular View: Standard atom-bond structure.
- Element View: Elemental knowledge (e.g., from the periodic table).
- Pharmacological/Drug View: External biological or functional knowledge [100].
This multi-perspective approach allows for a much richer and more nuanced understanding of molecular properties, which could be pivotal in interpreting the functional capabilities of molecules identified in ancient samples. The diagram below conceptualizes this integrative model.
Applications and Implications for Prebiotic Chemistry
The application of these ML-driven techniques has yielded dramatic results, pushing back the timeline of life and its metabolic processes. The detection of photosynthetic life in 2.5 billion-year-old rocks suggests that complex, light-harvesting biochemistry emerged nearly a billion years earlier than previously confirmed by molecular evidence [98] [96]. This provides a new temporal context for the environmental conditions in which later, more complex life evolved.
Furthermore, the ability to detect faint biosignatures in highly degraded samples offers a new pathway to test hypotheses about the RNA world. By analyzing the carbonaceous remains in ancient sedimentary rocks, researchers can now search for statistical evidence of patterns associated with RNA-like molecules or their breakdown products, even if the specific ancient compound cannot be fully resolved [97] [96]. This moves the investigation beyond the search for a single "smoking gun" molecule and towards a holistic analysis of the chemical context that implies a biological, and potentially prebiotic, origin.
Future Directions and Astrobiological Applications
The future of this field lies in refining these tools and expanding their applications. Key directions include:
- Dataset Expansion: To improve model accuracy, particularly for distinguishing between closely related biological sources, researchers are actively expanding their training sets, with a goal of over 1,000 analyzed samples [98].
- Tool Development for Complex Structures: Tools like MolParser, an end-to-end visual recognition system for molecule structures in scientific literature, will be crucial for automatically parsing and converting images of complex chemical structures into machine-readable formats, thus populating knowledge bases for analysis [101].
- Planetary Exploration: The methodology has direct applicability in the search for extraterrestrial life. NASA's Curiosity rover already carries a pyrolysis GC/MS instrument. In the future, a machine learning model could be deployed on a robotic mission to autonomously interpret data and identify potential biosignatures on Mars or other planetary bodies [98].
The fusion of advanced analytical chemistry with sophisticated machine learning is fundamentally transforming the study of life's origins. By providing a means to detect the faint, fragmented whispers of ancient biology, these techniques are building a novel, chemical record of life's early history on Earth. This new empirical evidence is critical for testing, refining, and validating theoretical frameworks like the RNA world hypothesis. As these tools continue to evolve, they will not only illuminate the deep past of our own planet but also guide humanity's search for life in the cosmos, providing a powerful, agnostic toolkit for recognizing life's signature, whatever form it may take.
Life, as we know it, is orchestrated by an intricate interplay between three fundamental molecular classes: deoxyribonucleic acid (DNA), ribonucleic acid (RNA), and proteins. This tripartite system forms the core of the central dogma of molecular biology, which posits that genetic information flows from DNA to RNA to proteins. DNA serves as the long-term repository of genetic information, RNA acts as a messenger and facilitator, and proteins execute the vast majority of cellular functions. However, the evolutionary origins of this sophisticated system remain a subject of intense scientific inquiry. The RNA World Hypothesis proposes that early life forms relied exclusively on RNA for both information storage and catalytic functions, predating the evolutionary emergence of DNA and proteins. This framework provides a critical lens through which to analyze the distinct properties, capabilities, and limitations of RNA, DNA, and proteins. Understanding their comparative strengths and weaknesses is essential not only for unraveling the history of life but also for advancing modern biomedical applications, including the development of RNA-based therapeutics and synthetic biological systems.
Structural and Functional Comparison of RNA, DNA, and Proteins
The distinct biological roles of nucleic acids and proteins are dictated by profound differences in their chemical structures, composition, and resultant stability.
Composition and Stability
Table 1: Fundamental Molecular Characteristics of DNA, RNA, and Proteins
| Characteristic | DNA | RNA | Proteins |
|---|---|---|---|
| Primary Function | Long-term genetic information storage [102] | Genetic information transfer, catalytic function (ribozymes) [102] [4] | Cellular structure, catalysis, signaling, regulation [103] |
| Sugar Component | Deoxyribose [102] | Ribose [102] | Not applicable |
| Key Bases | Adenine (A), Thymine (T), Guanine (G), Cytosine (C) [102] | Adenine (A), Uracil (U), Guanine (G), Cytosine (C) [102] | 20 different amino acids |
| Structural Form | Double-stranded helix [102] | Single-stranded (can form secondary structures) [102] | Complex 3D folding (primary, secondary, tertiary, quaternary) [103] |
| Stability | Highly stable due to deoxyribose sugar and double-stranded structure [102] | Less stable; reactive due to ribose sugar (2'-OH group) and single-stranded nature [102] [3] | Variable stability; dependent on amino acid sequence and environmental conditions [103] |
| Location in Eukaryotic Cell | Primarily nucleus [102] | Nucleolus and cytoplasm [102] | Throughout the cell |
The sugar backbone is a primary differentiator. DNA uses deoxyribose, which lacks a reactive hydroxyl group at its 2' carbon position, making the molecule less reactive and more stable. In contrast, RNA uses ribose, with a 2' hydroxyl group that renders the molecule more susceptible to hydrolysis and alkaline degradation [102]. This chemical difference has profound implications for molecular longevity, positioning DNA as a superior archive for genetic information and RNA as a transient mediator.
Regarding nitrogenous bases, both DNA and RNA utilize adenine, guanine, and cytosine. The key distinction lies in the fourth base: DNA employs thymine, while RNA uses uracil. In DNA, thymine's methyl group provides additional stability [102]. The base-pairing rules consequently differ, with DNA forming A-T and C-G pairs, and RNA forming A-U and C-G pairs.
Proteins, fundamentally different from nucleic acids, are polymers of amino acids linked by peptide bonds. Their identity and function are not determined by a linear sequence of bases but by the chemical properties of their amino acid side chains, which dictate folding into complex three-dimensional structures. This structural complexity underpins their diverse catalytic and structural roles [103].
Functional Versatility and the Case for an RNA World
The functional comparison of these molecules reveals why RNA is a compelling candidate for the primordial biopolymer.
Table 2: Functional Comparison and Support for the RNA World Hypothesis
| Aspect | DNA | RNA | Proteins |
|---|---|---|---|
| Information Storage | Excellent (high stability) [102] | Good (but less stable) [102] [3] | No inherent template-based replication |
| Catalytic Ability | Limited | Yes (as ribozymes) [4] [3] | Excellent (as enzymes) [103] |
| Self-Replication | Requires enzymatic machinery | Demonstrated in laboratory settings [4] | Not applicable |
| Evolutionary Capacity | Low (without proteins/RNA) | High (can evolve via mutation and selection) [4] | High (can evolve via mutation and selection) |
| Key Supporting Evidence for Primordial Role | Not considered a candidate for first life | Discovery of ribozymes; ribosome is a ribozyme [4] [3] | Cannot replicate or store genetic information independently |
RNA's dual capability is its defining feature. Like DNA, it can store genetic information through its nucleotide sequence. This is evidenced in many viruses that use RNA as their genetic material. Crucially, like proteins, certain RNA molecules—known as ribozymes—can fold into three-dimensional shapes and catalyze chemical reactions [4]. The most powerful evidence for the RNA World is the ribosome, the cellular machine that synthesizes proteins. The ribosome is a ribozyme; the catalytic activity that forms peptide bonds is performed by ribosomal RNA, not by the associated proteins [3]. This suggests that an RNA-based machinery was responsible for building the first proteins.
In contrast, DNA is a highly specialized but functionally limited information repository, lacking significant catalytic activity. Proteins, while functionally versatile and efficient catalysts, lack the ability to store heritable genetic information or self-replicate. Thus, RNA alone can perform both critical functions, supporting the hypothesis that it could have supported a primitive form of life before the advent of DNA and proteins [4].
The RNA World Hypothesis and Prebiotic Chemistry
The RNA World Hypothesis is a foundational concept in origins-of-life research, positing that there was a stage in early Earth's history where RNA both stored genetic information and catalyzed chemical reactions, preceding the evolutionary development of DNA-based genetics and protein-based catalysis [4] [3]. This theory, advanced in the 1960s by Carl Woese, Francis Crick, and Leslie Orgel, and later termed the "RNA World" by Walter Gilbert in 1986, suggests that around 4 billion years ago, RNA was the primary living substance [4]. The hypothesis resolves the "chicken-and-egg" dilemma of whether genetics or metabolism came first by proposing that a single molecule could fulfill both roles.
Key Evidence and Major Challenges
The discovery of ribozymes in the 1980s by Sidney Altman and Thomas Cech provided the first concrete evidence that RNA could indeed act as a catalyst, for which they received the Nobel Prize in Chemistry in 1989 [4] [3]. As previously noted, the central role of RNA in the ribosome's catalytic function is considered the strongest evidence for the hypothesis.
However, significant challenges remain. The prebiotic synthesis of RNA is chemically difficult. RNA nucleotides are complex molecules, and plausible pathways for their abiotic formation under early Earth conditions often produce complex, intractable mixtures with low yields of the desired components [104]. Furthermore, the RNA molecule itself is chemically fragile; its ribose-phosphate backbone is susceptible to hydrolysis, and it has a relatively limited range of catalytic activities compared to proteins [4] [3]. These limitations have led some researchers, like biochemist Harold S. Bernhardt, to critique the theory, pointing to the complexity and instability of RNA as major hurdles [3].
Modern Research: From "Pure" RNA to Systems Chemistry
In response to these challenges, the field of prebiotic chemistry is evolving. A newer approach, systems chemistry, moves away from the idea of a "pure" RNA world and instead explores how mixtures of different types of molecules could have worked together from the beginning. This perspective acknowledges that the transition from chemistry to life likely involved complex mixtures and synergistic interactions [105]. The goal is to find "Goldilocks chemistry"—reaction conditions on plausible prebiotic reactant mixtures that yield multiple biological building blocks in good yield without excessive by-products [105]. This includes exploring how the synthesis of nucleotides, amino acids, and lipids could have been compatible and even mutually reinforcing under common geochemical conditions [104] [105].
Diagram 1: Evolution of origins-of-life research from a pure "RNA World" to a "Systems Chemistry" perspective.
Experimental Approaches and Methodologies
Research in this field relies on innovative experimental designs to test the feasibility of prebiotic scenarios and to quantitatively analyze the relationships between RNA, DNA, and proteins in modern biological systems.
Simulating Prebiotic Environments with Microfluidic Devices
Microfluidic devices are small tools with channels on the micron scale that allow for the precise manipulation of very small fluid volumes. They are increasingly used in prebiotic chemistry to simulate the dynamic conditions of early Earth environments, such as hydrothermal vents [106]. These devices offer significant advantages, including reduced reagent consumption, high precision in mixing, and fast diffusive mixing, which allows for rapid testing of reaction conditions [106].
Three main types of microfluidic devices are used:
- Y-Form Devices with Laminar Co-Flow: Used to study mineral precipitation by bringing two fluids (e.g., simulating alkaline hydrothermal fluid and acidic ocean water) into contact without turbulent mixing, replicating conditions at hydrothermal vents [106].
- Microdroplet Devices: Generate tiny water-in-oil droplets that mimic cellular compartmentalization, allowing researchers to study how molecules can become concentrated and interact within enclosed spaces [106].
- Devices with Microchambers: Recreate the microscopic pores within rocks on the seafloor, providing a confined environment for chemical reactions to occur [106].
Correlating RNA and Protein Expression
A key tenet of the Central Dogma is that DNA makes RNA makes protein. However, the relationship between RNA abundance (mRNA) and protein abundance is not straightforward. A large-scale study analyzing 1,066 genes across 23 human cell lines found that the mean correlation between mRNA and protein levels was only 0.20-0.25 [107]. This indicates that for many genes, mRNA levels are a poor predictor of protein abundance, highlighting the significant role of post-transcriptional regulation (e.g., translation efficiency, protein degradation).
More recent, powerful studies using single-cell analysis in yeast have reinforced this, showing that less than 20% of genetic loci that influence gene expression have concordant effects on both mRNA and protein levels for the same gene. The majority specifically affect protein levels without altering mRNA abundance [108]. This complexity underscores the sophisticated multi-layered regulation that has evolved beyond a simple RNA-centric system.
Simultaneous Biomolecule Extraction for Integrated Analysis
To comprehensively study molecular systems, researchers have developed protocols for the simultaneous extraction of DNA, RNA, proteins, and metabolites from a single, limited biological sample. This approach is crucial for "omics" studies that aim to integrate genomics, transcriptomics, proteomics, and metabolomics. Optimized methods involve a step of methanol/chloroform purification for metabolites before the separation of DNA/RNA and proteins, ensuring all biomolecule classes are obtained in adequate quantity and quality for downstream analysis [109].
The Scientist's Toolkit: Key Research Reagents and Methods
Table 3: Essential Reagents and Methods for RNA World and Prebiotic Research
| Reagent / Method | Function in Research | Relevance to RNA World & Comparative Analysis |
|---|---|---|
| Ribozymes | Catalytic RNA molecules used to demonstrate RNA's enzymatic potential. | Key evidence for the hypothesis; used to study prebiotic catalysis and RNA evolution [4] [3]. |
| Microfluidic Devices | Lab-on-a-chip systems to simulate prebiotic microenvironments. | Used to study chemical gradients, compartmentalization, and reactions under plausible early Earth conditions [106]. |
| Formamide & HCN Chemistry | Simple C1 feedstock molecules for prebiotic synthesis. | Starting points for generating nucleobases, nucleotides, and amino acids in simulated prebiotic reactions [104]. |
| Simultaneous Extraction Kits | Kits for co-extracting DNA, RNA, proteins, and metabolites. | Enable integrated multi-omics analysis to study relationships between different molecular classes in modern systems [109]. |
| Dual Fluorescent Reporters | Genetically engineered systems for simultaneous mRNA and protein quantification in single cells. | Allow precise correlation of transcriptional and translational regulation, revealing post-transcriptional control [108]. |
Diagram 2: An integrated experimental workflow for the comparative analysis of DNA, RNA, and protein expression in modern biological systems.
The comparative analysis of RNA, DNA, and protein-based systems reveals a compelling evolutionary narrative. RNA's unique combination of genetic and catalytic functions solidifies its central role in the RNA World Hypothesis as the likely cornerstone of early life. While challenges regarding its prebiotic synthesis and stability persist, they are driving innovative research in systems chemistry that explores how interconnected networks of molecules could have given rise to life. The evolutionary transition to a tripartite system of DNA, RNA, and proteins allowed for a division of labor, leveraging the superior stability of DNA for information storage and the superior catalytic efficiency of proteins for cellular operations. Modern experimental techniques, from microfluidics to single-cell omics, continue to refine our understanding of these molecules' distinct yet interconnected roles. This knowledge not only illuminates the deep past but also informs the future of biotechnology, where engineered RNA molecules and synthetic protocells stand as testaments to the enduring versatility of this primordial biopolymer.
The RNA World Hypothesis proposes that earlier life forms may have relied solely on RNA for both storing genetic information and catalyzing essential chemical reactions, prior to the evolution of DNA and proteins [4] [110]. This concept, first advanced in the 1960s by Carl Woese, Francis Crick, and Leslie Orgel, suggests that around 4 billion years ago, RNA was the primary living substance because of its dual capabilities [4] [3]. The central reasoning is that RNA is capable of self-replication, and could therefore have carried genetic information across generations independently [4]. The hypothesis gained significant support with the discovery of ribozymes—RNA molecules capable of catalysis—by Sidney Altman and Thomas Cech, for which they received the Nobel Prize in Chemistry in 1989 [4] [3].
The ultimate goal of laboratory recreation is to mimic these proposed early evolutionary steps by constructing autonomous RNA-based life from simple molecular building blocks. As Senior author Gerald Joyce of the Salk Institute states, "We’re chasing the dawn of evolution... By revealing these novel capabilities of RNA, we’re uncovering the potential origins of life itself, and how simple molecules could have paved the way for the complexity and diversity of life we see today" [44]. This whitepaper details the technical methodologies and experimental protocols driving this frontier of prebiotic chemistry research.
Foundational Principles and Recent Breakthroughs
The Case for RNA as the Pioneer Biopolymer
RNA's suitability as a candidate for the first self-replicating molecule stems from its core properties [110] [44] [3]:
- Informational Capacity: Like DNA, RNA can store genetic information in its nucleotide sequence.
- Catalytic Function: As a ribozyme, RNA can catalyze chemical reactions, including those critical for self-replication.
- Structural Flexibility: Single-stranded RNA folds into complex three-dimensional structures, enabling diverse functions.
This combination of capabilities makes RNA uniquely suited to have initiated Darwinian evolution before the biological division of labor among DNA, RNA, and proteins [44].
Key Experimental Validation
Recent research has demonstrated RNA's capacity for sustained evolution. A pivotal 2024 study published in Proceedings of the National Academy of Sciences revealed an RNA enzyme capable of replicating RNA strands accurately while allowing for the emergence of new molecular variants over time [44]. This research created RNA polymerase ribozymes with crucial mutations that enhance replication accuracy—a fundamental requirement for evolution. The study also documented a "hammerhead" RNA molecule capable of both self-replication and variation, demonstrating a simple yet effective form of early evolution where molecular-level changes could spark biological complexity [44].
First author Nikolaos Papastavrou reflects on the implications: "We’ve long wondered how simple life was at its beginning and when it gained the ability to start improving itself. This study suggests the dawn of evolution could have been very early and very simple. Something at the level of individual molecules could sustain Darwinian evolution, and that might have been the spark that allowed life to become more complex, going from molecules to cells to multicellular organisms" [44].
Experimental Protocols for RNA Synthesis and Evolution
Enzymatic Synthesis of RNA Polymers
The synthesis of RNA molecules for experimentation employs both well-established and novel enzymatic methods.
Standard In Vitro Transcription [111]: This routine procedure allows template-directed synthesis of RNA molecules of any sequence from short oligonucleotides to several kilobases. The protocol is based on engineering a DNA template that includes a bacteriophage promoter sequence (e.g., from T7 phage) upstream of the sequence of interest, followed by transcription using the corresponding RNA polymerase.
Table 1: Key Components for Standard In Vitro Transcription
| Component | Function | Considerations |
|---|---|---|
| DNA Template | Contains promoter sequence and desired RNA sequence | Must include bacteriophage promoter (e.g., T7, SP6, T3) |
| RNA Polymerase | Catalyzes RNA synthesis from DNA template | T7 RNA polymerase commonly used for high yield |
| Ribonucleotides | Building blocks for RNA synthesis (ATP, UTP, GTP, CTP) | Typically used at 1-2 mM each; often includes cap analog for mRNA |
| Reaction Buffer | Provides optimal ionic conditions | Typically contains Mg²⁺, DTT, salts; varies with polymerase |
Novel Enzymatic Synthesis for Random Libraries [112]: For generating large, diverse RNA libraries—particularly those with random sequences for aptamer or ribozyme selection—researchers have developed an enzymatic method using human DNA polymerase Theta (θ) mutants. These mutants can generate long single-stranded RNA polynucleotides of random sequences due to their improved template-free terminal nucleotidyltransferase activity. This protocol enables the rapid synthesis of RNA polymers thanks to the efficient incorporation of ribonucleotides as well as chemically modified ribonucleotides, producing libraries ready for repeated cycles of Systematic Evolution of Ligands by Exponential enrichment (SELEX) [112].
Directed Evolution of Functional RNA
The process of evolving RNA molecules with enhanced functions follows an iterative selection and amplification protocol:
Table 2: Key Steps in RNA Directed Evolution
| Step | Process | Outcome |
|---|---|---|
| Library Generation | Create large, diverse pool of RNA sequences | 10¹³-10¹⁵ different sequences for selection |
| Selection Pressure | Apply conditions that favor desired function | Enrichment of functional sequences |
| Amplification | Reverse transcribe, PCR amplify selected RNA | Increase copy number of selected variants |
| Transcription | Generate RNA for next selection cycle | New RNA pool with enriched functionality |
| Iteration | Repeat selection/amplification cycles | Progressive improvement of function |
Co-author David Horning notes the ongoing research questions: "We've seen that selection pressure can improve RNAs with an existing function, but if we let the system evolve for longer with larger populations of RNA molecules, can new functions be invented? We're excited to answer how early life could ratchet up its own complexity, using the tools developed here at Salk" [44].
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful recreation of autonomous RNA systems requires specific, high-quality materials and reagents.
Table 3: Essential Research Reagents for RNA World Experiments
| Reagent/Material | Function/Application | Technical Notes |
|---|---|---|
| Ribonucleotide Triphosphates | Building blocks for RNA synthesis | High-purity NTPs reduce abortive transcription; can include modified bases |
| T7 RNA Polymerase | Workhorse enzyme for in vitro transcription | High-yield synthesis; commercially available recombinant forms |
| DNA Polymerase Theta Mutants | Generating random RNA libraries | Template-free activity for diverse library generation [112] |
| Ribozyme Scaffolds | Starting points for evolution studies | Hammerhead, hairpin, and other self-cleaving motifs |
| Homogeneity Reference Materials | Quality control and standardization | Certified reference materials like Quartet RNA for data reliability [113] |
| Modified Ribonucleotides | Enhancing nuclease resistance | C2'-modified nucleotides (e.g., 2'-O-methyl) for stability [112] |
Pathway to Autonomous RNA Life: Experimental Workflow
The comprehensive approach to building autonomous RNA-based life involves multiple interconnected experimental phases, from initial synthesis to the emergence of evolutionary capacity.
Current Challenges and Research Directions
Despite significant advances, researchers face several substantial challenges in recreating autonomous RNA-based life:
- Prebiotic Plausibility: The complexity of RNA molecules raises questions about how they could have arisen prebiotically from simple non-living chemicals [4] [3].
- Chemical Instability: RNA is relatively unstable compared to DNA, particularly due to the presence of the 2'-hydroxyl group which makes it more prone to hydrolysis [4] [110].
- Limited Catalytic Range: While ribozymes exist, they generally possess narrower catalytic activities compared to protein enzymes [4].
NASA noted in a 1996 report that "significant difficulties" surrounding the RNA World concept include RNA's chemical fragility and its narrow range of catalytic activities [4]. Biochemist Harold Bernhardt has similarly expressed concerns, noting in a 2012 paper that the complexity of RNA means it could not have arisen prebiotically [4].
Future research aims to address these challenges by exploring environmental conditions conducive to RNA evolution, developing RNA molecules with enhanced replicative and catalytic abilities, and potentially extending this inquiry to the potential for life beyond Earth [44]. The Salk Institute team believes that recreating RNA-based life in the laboratory is achievable within the next decade [44].
The experimental recreation of autonomous RNA-based life from simple molecules represents one of the most ambitious goals at the intersection of prebiotic chemistry and synthetic biology. By demonstrating that RNA can replicate, evolve, and potentially increase in complexity under laboratory conditions, researchers are not only testing the RNA World Hypothesis but also pioneering new approaches to understanding life's fundamental principles.
The protocols and methodologies detailed in this whitepaper provide a roadmap for researchers pursuing this frontier. As these experimental techniques refine our ability to generate, evolve, and characterize RNA molecules, we move closer to answering one of humanity's most profound questions: How did life begin? The implications extend beyond origins of life research to synthetic biology, biotechnology, and even the search for extraterrestrial life [44].
A groundbreaking study employing machine learning to analyze molecular fragments in ancient rocks has identified chemical evidence of oxygen-producing photosynthesis in 2.5-billion-year-old samples. This finding pushes back the molecular record of this pivotal metabolic process by nearly 800 million years, offering profound insights into the early evolution of life on Earth. The research, which pairs pyrolysis gas chromatography-mass spectrometry (GC/MS) with artificial intelligence, demonstrates a novel capability to detect degraded biosignatures, thereby extending the temporal limits of the geological record and providing a powerful new tool for probing the interfaces of prebiotic chemistry and the RNA world hypothesis.
Understanding the emergence of life and its early metabolic capabilities represents one of the most significant challenges in science. For most of Earth's 4.5-billion-year history, life was exclusively microbial [114]. The fossil record indicates that microbial mats and stromatolites—layered structures formed by cyanobacteria and other microorganisms—were widespread by 3.5 billion years ago [115] [116]. However, direct molecular evidence for specific metabolic processes, particularly photosynthesis, has been largely confined to rocks younger than 1.7 billion years due to the degradation of biomolecules over geological time [117].
The context for life's origin is often framed by the RNA world hypothesis, which posits that early life was based on RNA molecules capable of both storing genetic information and catalyzing chemical reactions, predating the DNA-protein world [23] [44]. This hypothesis suggests that RNA, or similar nucleic acids, were central players in prebiotic chemistry, eventually giving rise to self-replicating systems and the first cellular life [52]. Within this framework, the emergence of photosynthesis—a process that ultimately transformed Earth's atmosphere—marks a critical evolutionary transition from primitive heterotrophic or chemosynthetic organisms to phototrophic life that harnessed solar energy.
A New Analytical Paradigm: Machine Learning and Chemical 'Whispers'
Experimental Protocol and Workflow
The recent breakthrough in detecting ancient photosynthesis stems from a innovative methodology that combines advanced analytical chemistry with machine learning [118] [117]. The research team analyzed 406 diverse samples to train a model capable of distinguishing biological from non-biological materials even in highly degraded states.
Table 1: Sample Categories Used for Machine Learning Training
| Sample Category | Examples | Number of Samples |
|---|---|---|
| Modern Animals | Fish, insects | Not Specified |
| Modern Plants | Leaves, roots, sap | Not Specified |
| Fungi | Mushrooms, yeast | Not Specified |
| Fossil Materials | Coal, ancient wood, algae-rich shale | 406 total across all categories |
| Meteorites | Carbon-rich space rocks | Not Specified |
| Synthetic Organics | Lab-simulated early-Earth chemistry | Not Specified |
| Ancient Sediments | Rocks from hundreds of millions to over 3 billion years old | Not Specified |
The core experimental protocol followed these key steps:
- Sample Pyrolysis and GC/MS Analysis: Each sample was subjected to pyrolysis gas chromatography-mass spectrometry (GC/MS). This technique heats samples to high temperatures in an oxygen-free environment, breaking them down into smaller molecular fragments that are then separated by gas chromatography and identified by mass spectrometry [98] [117].
- Machine Learning Classification: The resulting complex fragmentation data from all samples were used to train a "random forest" machine learning model. This type of model builds hundreds of decision trees to classify data based on learned patterns [117].
- Pattern Recognition vs. Specific Biomarkers: Rather than targeting specific, known biomarker molecules that degrade over time, the model was trained to recognize the overall "chemical fingerprint" or pattern of fragments that is characteristic of biological origin, even when the original biomolecules are gone [119] [117]. As researcher Robert Hazen analogized, "Think of it like showing thousands of jigsaw puzzle pieces to a computer and asking whether the original scene was a flower or a meteorite" [117].
Model Performance and Validation
The trained model demonstrated a remarkable ability to discern the origin of organic matter, achieving the following accuracy rates [117]:
- Distinguishing biological from non-biological materials: Up to 98% accuracy.
- Identifying photosynthetic signatures: 93% accuracy.
- Differentiating plant-based from animal-based life: 95% accuracy.
The model's performance was validated using samples with known origins before being applied to ancient, unknown samples. This high level of accuracy confirmed that the "chemical echoes" of life persist in molecular fragments long after definitive biomarkers have degraded [119].
Groundbreaking Findings: Pushing Back the Timeline of Photosynthesis
The application of this new analytical technique has yielded two transformative findings regarding the early history of life.
Extending the Molecular Record of Life
The machine learning model detected clear chemical evidence of life in 3.3-billion-year-old rocks from the Josefsdal Chert formation in South Africa [117]. This finding is significant because it effectively doubles the window of time in which organic molecules can reveal information about ancient life. Prior to this study, the oldest molecular traces that could be confidently linked to biology came from rocks no older than 1.7 billion years [117].
The Earliest Evidence of Oxygenic Photosynthesis
Furthermore, the model identified molecular signatures of oxygen-producing photosynthesis in 2.52-billion-year-old rocks from the Gamohaan Formation in South Africa [118] [117]. This pushes back the molecular evidence for photosynthesis by approximately 800 million years relative to the previous chemical record. This timing is critical as it precedes the Great Oxygenation Event (GOE), which began around 2.4 billion years ago—a period when Earth's atmosphere experienced a significant rise in oxygen levels largely attributed to photosynthetic microorganisms [118].
Table 2: Key Findings from Ancient Rock Analysis
| Rock Formation | Age (Billion Years) | Key Finding | Scientific Significance |
|---|---|---|---|
| Josefsdal Chert (South Africa) | 3.3 | Chemical evidence of life | Doubles the previous molecular record of life (previously 1.7 billion years) |
| Gamohaan Formation (South Africa) | 2.52 | Molecular signatures of oxygenic photosynthesis | Pushes back chemical evidence of photosynthesis by ~800 million years; precedes the Great Oxygenation Event |
Implications for Prebiotic Chemistry and the RNA World
The discovery of sophisticated photosynthetic life at 2.5 billion years ago has profound implications for understanding the earlier phases of life's evolution, stretching back into the era of prebiotic chemistry and the hypothesized RNA world.
Compressing the Evolutionary Timeline
The existence of oxygenic photosynthesis 2.5 billion years ago implies that the preceding evolutionary steps must have occurred even earlier. The RNA world, if it existed, would have dominated the pre-2.5 billion-year landscape, with its transition to a DNA/protein world and the development of complex metabolic pathways like photosynthesis requiring substantial time [23] [44]. This new evidence places tighter constraints on the timing of these major evolutionary transitions, suggesting a relatively rapid emergence of biological complexity from prebiotic beginnings.
The Role of Non-Canonical Nucleotides
Recent research into prebiotic evolution has highlighted the potential importance of non-canonical nucleotides (beyond the standard A, U, G, C) in the early stages of life's development. It has been shown that different forms of RNA can include about 170 such non-canonical nucleotides, which may have played crucial roles in the emergence of catalytic RNAs and the origin of template-directed synthesis [52]. The complexity inherent in a photosynthetic system at 2.5 billion years ago suggests a long prior history of molecular evolution involving these diverse molecular building blocks.
The Scientist's Toolkit: Key Research Reagents and Materials
The experimental breakthroughs described herein rely on a suite of sophisticated analytical tools and reagents. The following table details key solutions and materials central to this research paradigm.
Table 3: Essential Research Reagents and Analytical Solutions
| Reagent / Material | Function / Application | Experimental Role |
|---|---|---|
| Pyrolysis GC/MS System | Analytical instrument suite for fragmenting samples and characterizing molecular pieces. | Core platform for generating the complex chemical fragmentation data used for machine learning analysis [98] [117]. |
| Certified Reference Materials (CRMs) | Pure organic compounds and abiotic carbon samples (e.g., meteorites, synthetic organics) with known composition. | Serves as control and training set for the machine learning model, enabling it to learn the difference between biotic and abiotic chemical patterns [117]. |
| Ancient Sediment Samples | Crushed rock powders from well-characterized geological formations (e.g., Josefsdal Chert, Gamohaan Formation). | The primary unknown samples under investigation; source of the degraded biosignatures from deep time [117]. |
| Random Forest Algorithm | A machine learning model composed of many decision trees. | The classification engine that identifies subtle, complex patterns in the GC/MS fragmentation data that are indicative of biological origin or specific metabolisms [117]. |
Discussion and Future Directions
The ability to detect chemical "whispers" of life in billion-year-old rocks represents a paradigm shift in paleobiology. This technique moves beyond the search for specific, intact biomarker molecules—which are easily lost to geological processes—and instead leverages the persistent statistical patterns in molecular fragments [119] [117]. This approach is particularly powerful for resolving long-standing debates about the nature of Earth's earliest life, as it can be applied to the vast majority of ancient carbon-bearing rocks that preserve neither visible fossils nor pristine biomolecules.
A critical future direction will be to apply this method to older rocks, particularly those from the Archean Eon (4.0 to 2.5 billion years ago), to search for the chemical footprints of even more primitive life forms, potentially including anoxygenic photosynthetic bacteria or the last universal common ancestor (LUCA). Furthermore, the success of this methodology has immediate implications for astrobiology. NASA rovers like Curiosity already carry pyrolysis-GC/MS instruments [98]. Implementing similar machine learning algorithms could enable robotic missions to identify potential biosignatures on Mars or other planetary bodies, even if they represent alien biochemical systems not based on Earth-like biology [118] [117].
The detection of photosynthetic signatures in 2.5-billion-year-old rocks through machine learning analysis of molecular fragments provides a new, powerful lens through which to view the dawn of life. This evidence firmly establishes that oxygenic photosynthesis—a process that would ultimately remake Earth's atmosphere—was active nearly a billion years earlier than the molecular record previously showed. By bridging the gap between the theoretical RNA world and the tangible geological record, this research provides a more constrained and detailed timeline for the rise of biological complexity. The fusion of advanced analytical chemistry with artificial intelligence not only illuminates the deep past on Earth but also provides an indispensable tool for seeking signs of life beyond our planet.
The RNA World hypothesis represents a dominant paradigm in origins of life research, proposing that early life on Earth was based on RNA molecules that served both genetic and catalytic functions prior to the evolutionary emergence of DNA and proteins [23]. This hypothesis challenges the traditional view that these macromolecules were fundamental components from life's inception, instead positioning RNA as a pivotal precursor that could store genetic information while simultaneously catalyzing the chemical reactions necessary for life [23]. Despite its explanatory power, the RNA World hypothesis faces significant challenges, particularly regarding the prebiotic synthesis of RNA nucleotides and the inherent instability of RNA molecules under early Earth conditions [23] [84]. These difficulties have prompted researchers to investigate whether RNA itself might have been preceded by simpler, more robust genetic systems.
This investigation has given rise to the concept of a Pre-RNA World, which explores the possibility that earlier genetic materials may have paved the way for RNA's eventual dominance [120] [84]. Within this theoretical framework, several nucleic acid analogs have emerged as compelling candidates for pre-RNA genetic systems. Peptide Nucleic Acid (PNA), Threose Nucleic Acid (TNA), and Glycerol Nucleic Acid (GNA) constitute the most prominent subjects of current research due to their structural simplicity, chemical robustness, and capacity for information storage [121] [120] [122]. These alternative genetic systems potentially resolve key limitations of RNA in prebiotic contexts, offering plausible pathways for the emergence of molecular evolution before the RNA World. This whitepaper provides a comprehensive technical analysis of PNA, TNA, and GNA as potential pre-RNA candidates, examining their structural properties, experimental support, and implications for understanding life's chemical origins.
Structural Properties and Comparative Analysis of Pre-RNA Candidates
The structural architectures of PNA, TNA, and GNA differ fundamentally from the ribose-phosphate backbone of canonical RNA, conferring distinct chemical properties that may have been advantageous in prebiotic environments. Understanding these molecular designs is crucial for evaluating their feasibility as primordial genetic materials.
Peptide Nucleic Acid (PNA): PNA features a backbone where the entire sugar-phosphate structure of RNA is replaced by N-(2-aminoethyl)glycine units linked by peptide bonds [121]. The various purine and pyrimidine bases are attached to this backbone via a methylene bridge (-CH₂-) and a carbonyl group (-(C=O)-) [121]. This architecture results in an achiral, uncharged molecule that lacks the negative charges inherent in natural nucleic acid backbones, enabling strong hybridization affinity to complementary DNA and RNA sequences due to the absence of electrostatic repulsion [121].
Threose Nucleic Acid (TNA): TNA utilizes the four-carbon sugar threose instead of the five-carbon ribose found in RNA, creating a structurally simplified genetic system [120]. The threose sugar is smaller than ribose, potentially making TNA easier to form under prebiotic conditions [120]. Despite this simplification, TNA can form stable Watson-Crick antiparallel duplex structures with itself and with RNA, providing a mechanism for genetic information transfer between successive genetic systems [123] [120].
Glycerol Nucleic Acid (GNA): GNA represents an even more minimalist architecture with an acyclic backbone derived from three-carbon glycerol units connected via phosphodiester bonds [122]. This structure contains a single chiral center and serves as an isostere of RNA, maintaining similar bonding patterns while offering greater conformational flexibility [122]. GNA can form stable helical duplexes and has demonstrated particular utility in therapeutic applications, such as improving the safety profile of RNAi therapeutics [122].
Table 1: Comparative Structural Properties of Pre-RNA Candidate Molecules
| Property | PNA | TNA | GNA |
|---|---|---|---|
| Backbone Composition | N-(2-aminoethyl)glycine | Threose-phosphate | Glycerol-phosphate |
| Chirality | Achiral | Chiral | Chiral (single center) |
| Charge | Neutral | Negative | Negative |
| Base Pairing | Watson-Crick & Hoogsteen | Watson-Crick | Reverse Watson-Crick |
| Thermal Stability | High (Tm ~70°C for 15mer) | Moderate | Varies with sequence |
| Nuclease Resistance | High | Not specified | High |
The following diagram illustrates the structural evolution from potential pre-RNA genetic systems to the contemporary DNA/RNA-based biology:
Structural Evolution to RNA World
These structural properties have profound implications for the prebiotic feasibility of each candidate. PNA's neutrality and chemical robustness [121], TNA's simplified sugar backbone [120], and GNA's minimalistic acyclic structure [122] each address different limitations of RNA in prebiotic contexts, potentially enabling molecular evolution under early Earth conditions.
Experimental Evidence and Research Methodologies
Key Experimental Findings
Rigorous laboratory investigations have provided critical insights into the functional capabilities of pre-RNA candidates, testing their potential to serve as genetic materials prior to the emergence of RNA.
PNA Research Methodologies and Findings PNA studies often employ solid-phase synthesis techniques adapted from peptide chemistry, allowing for the production of specific PNA oligomers [121]. Research has demonstrated that PNA can form highly stable duplex invasion complexes with double-stranded DNA, particularly through bis-PNA structures where Watson-Crick and Hoogsteen binding domains are connected via flexible linkers [121]. Thermal denaturation studies reveal exceptionally high melting temperatures (Tm ~70°C for a 15mer PNA-DNA duplex), with significantly greater discrimination against mismatched bases (ΔTm = 15°C for single mismatch in PNA-DNA vs. 11°C for DNA-DNA) [121]. PNA exhibits remarkable resistance to enzymatic degradation by nucleases and proteases, maintaining integrity for over 48 hours in cellular environments compared to less than 15 minutes for unmodified DNA/RNA oligonucleotides [121].
TNA Experimental Approaches and Results TNA research utilizes in vitro evolution methodologies to explore functional capabilities. In seminal experiments, researchers created TNA libraries and evolved them in the presence of target proteins [120]. Within just three generations, they isolated TNA molecules capable of folding into complex three-dimensional shapes and binding specifically to target proteins - key steps toward catalytic function [120]. Additional investigations employing thermal denaturation experiments have demonstrated that TNA can form stable helical duplexes with complementary TNA strands and with RNA, facilitating potential genetic information transfer [123]. However, these studies also revealed that GNA and TNA mixed sequence polymers cannot form stable helical structures through intersystem cross-pairing, suggesting they were not consecutive polymers in the same evolutionary pathway to RNA [123].
GNA Research Techniques and Outcomes GNA studies often utilize chemical synthesis approaches combined with structural analysis techniques like X-ray crystallography [122]. These investigations have revealed that GNA nucleotides adopt a rotated nucleobase orientation within duplex structures, pairing with complementary RNA in a reverse Watson-Crick mode [122]. This distinctive pairing geometry explains the inability of standard GNA C and G nucleotides to form strong base pairs with complementary RNA nucleotides. Researchers have addressed this limitation by developing novel (S)-GNA isocytidine and isoguanosine nucleotides with transposed hydrogen bond donor and acceptor patterns, successfully creating stable base-pairing systems with complementary ribonucleotides [122].
Table 2: Experimental Evidence Supporting Pre-RNA Candidate Functions
| Experimental Method | PNA Findings | TNA Findings | GNA Findings |
|---|---|---|---|
| Thermal Denaturation | High Tm values; Strong mismatch discrimination | Stable duplex formation with RNA | Stable pairing with modified bases |
| In vitro Evolution | Not applicable | Protein-binding molecules in 3 generations | Not specified |
| Structural Analysis | Duplex invasion complexes | Antiparallel duplex geometry | Reverse Watson-Crick pairing |
| Enzymatic Stability | Resistant to nucleases/proteases (>48h) | Not specified | Increased exonuclease resistance |
| Polymerization Feasibility | Possible at 100°C | Simplified precursor synthesis | Acyclic backbone synthesis |
The following diagram illustrates a generalized experimental workflow for evaluating pre-RNA candidates in origins of life research:
Pre-RNA Candidate Evaluation Workflow
The Scientist's Toolkit: Essential Research Reagents and Materials
Research into pre-RNA candidates requires specialized reagents and methodologies tailored to the unique chemical properties of these synthetic genetic systems.
Table 3: Essential Research Reagents for Pre-RNA Candidate Investigations
| Reagent/Material | Function | Application Examples |
|---|---|---|
| N-(2-aminoethyl)glycine monomers | PNA backbone synthesis | Solid-phase PNA oligomer assembly |
| Threose nucleosides | TNA building blocks | TNA strand synthesis and polymerization |
| Glycerol-derived phosphoramidites | GNA monomer units | GNA oligonucleotide synthesis |
| Diamidophosphate (DAP) | Prebiotic phosphorylation agent | Sugar phosphorylation studies [25] |
| Montmorillonite clay | Mineral catalyst | Template-directed polymerization [84] |
| Bis-PNA linkers | Bivalent PNA constructs | Duplex invasion complex formation [121] |
| Modified nucleotides (isoguanosine, isocytidine) | Alternative base pairing | GNA-RNA hybridization studies [122] |
Research Implications and Future Directions
Implications for the RNA World Hypothesis
Experimental investigations of PNA, TNA, and GNA have profound implications for our understanding of molecular evolution and the possible transition from a pre-RNA world to the RNA world. The demonstrated capacity of these alternative genetic systems to store information and potentially evolve [120] supports a modular transition hypothesis in which multiple genetic systems may have coexisted before RNA achieved dominance. However, research indicating that GNA and TNA cannot form stable intersystem helical structures [123] suggests these molecules may represent alternative evolutionary pathways rather than sequential predecessors to RNA.
The structural and functional properties of pre-RNA candidates provide plausible solutions to significant challenges in RNA-first scenarios. PNA's extreme robustness and potential for spontaneous polymerization at elevated temperatures [121] address concerns about RNA stability under prebiotic conditions. TNA's simplified threose backbone [120] offers a chemically accessible route to genetic polymer formation. GNA's minimalistic acyclic structure [122] demonstrates that even highly simplified nucleic acid analogs can support information storage and transfer. These findings collectively suggest that the emergence of genetic systems may have been facilitated by a diversity of molecular solutions rather than a single predetermined path.
Therapeutic Applications and Biotechnology
Beyond origins of life research, pre-RNA candidates have found practical applications in biotechnology and medicine, particularly in molecular diagnostics and therapeutic development. PNA's high binding affinity, sequence specificity, and resistance to enzymatic degradation have made it valuable for antisense applications, gene regulation strategies, and molecular diagnostics where discrimination of single-nucleotide polymorphisms is essential [121]. The neutral PNA backbone enables hybridization under low ionic strength conditions where DNA and RNA probes would be ineffective [121].
GNA modifications have demonstrated significant utility in RNAi therapeutics, where incorporation of (S)-GNA nucleotides into siRNAs improves safety profiles by mitigating off-target effects while maintaining gene silencing potency [122]. These modifications increase resistance to 3'-exonuclease-mediated degradation and enhance the therapeutic index of RNA-based medicines [122]. Clinical development of GNA-modified siRNAs represents a direct practical application of pre-RNA research to human therapeutics [122].
Future Research Trajectories
Several promising research directions emerge from current understanding of pre-RNA candidates. A primary focus involves establishing continuous experimental pathways from simple prebiotic precursors to functional polymers under plausible early Earth conditions [124] [125]. This includes investigating how nucleotide synthesis might align with protometabolic pathways, particularly those centered on reductive Krebs cycle intermediates and hydrothermal vent chemistry [124]. The discovery that ribose is selectively phosphorylated from mixtures of prebiotic sugars [25] provides a template for such investigations.
Additional frontiers include expanding in vitro evolution methodologies to explore the catalytic potential of TNA, GNA, and PNA systems more comprehensively [120], and developing computational models of alternative genetic systems to predict their evolutionary dynamics. The recent finding that contemporary RNA incorporates numerous non-canonical nucleotides [52] [125] suggests these modifications may represent molecular fossils from earlier evolutionary stages, providing another rich avenue for investigation into the transition between genetic systems.
The experimental investigation of PNA, TNA, and GNA as potential pre-RNA candidates has substantially enriched our understanding of molecular evolution and the possible pathways to life's emergence. Rather than supporting a simple linear progression from one genetic system to another, research reveals a complex landscape of molecular possibilities in which multiple nucleic acid analogs may have coexisted or competed before biological evolution settled on the RNA-DNA-protein system that characterizes contemporary life [120]. This perspective transforms the "RNA World" from a singular event into a potential evolutionary endpoint of a more extensive chemical evolutionary process.
The structural and functional properties of pre-RNA candidates demonstrate that genetic information storage and evolution need not be exclusive to RNA, offering plausible solutions to significant challenges in prebiotic chemistry. As research continues to bridge the gap between prebiotic chemistry and early biological evolution, these alternative genetic systems provide powerful experimental models for testing hypotheses about life's earliest stages. Their investigation not only illuminates life's possible origins on Earth but also expands the conceptual space for considering what forms life might take elsewhere in the universe.
The ribosome, an intricate ribonucleoprotein complex central to translation, functions as a molecular archive preserving records of its ancient evolution. This whitepaper examines the principle of evolutionary accretion through which the ribosome gained its modern layered architecture. We analyze timelines of structural accretion derived from computational and structural biology studies, revealing how a primitive proto-ribosome expanded through the coordinated addition of RNA and protein components. Within the framework of the RNA world hypothesis and prebiotic chemistry, we trace the co-evolution of ribosomal proteins and RNA, highlighting the transition from a primordial ribozyme to a sophisticated allosteric machinery. The analysis underscores how this accretion process facilitated the development of functional centers for decoding, peptidyl transfer, and allosteric communication, with significant implications for understanding the origins of biological complexity and guiding synthetic biology efforts.
The modern ribosome presents a complex structure universally conserved across all domains of life, indicating that its core architecture predates the Last Universal Common Ancestor (LUCA) [126]. Its historical layers provide a unique window into early evolutionary processes. The "accretion" hypothesis posits that the ribosome did not emerge in its fully-fledged form but rather grew outward from an ancient catalytic core through the sequential addition of structural subunits [127] [128]. This process involved the continuous integration of new RNA segments and ribosomal proteins (r-proteins), creating a stratified record of molecular evolution.
This structural narrative is deeply intertwined with the RNA world hypothesis, which suggests that early life was based primarily on RNA catalysis before the advent of coded protein synthesis [129] [130]. Within this framework, the ribosome's peptidyl transferase center (PTC) is considered a molecular fossil—a relic of an ancient ribozyme that catalyzed peptide bond formation in a primitive peptide world [129] [126]. The subsequent layering of RNA and protein components around this center enhanced both the fidelity and efficiency of translation, eventually leading to the sophisticated machinery observed today. This whitepaper synthesizes findings from phylogenetic, structural, and computational studies to trace the ribosome's accretional history and its implications for prebiotic chemistry.
The Proto-Ribosome and Prebiotic Foundations
The journey of the ribosome begins with the spontaneous emergence of a catalytic RNA core capable of facilitating peptide bond formation. Probabilistic analyses suggest that the simplest viable proto-ribosome likely consisted of a dimer of tRNA-like molecules embedded within the contemporary ribosome's symmetrical region [129]. This simple structure represents the only configuration with a realistic statistical likelihood of spontaneous emergence from random RNA chains in the prebiotic environment [129].
Table 1: Proto-Ribosome Structural Candidates and Their Characteristics
| Structural Element | Structural Complexity | Probability of Spontaneous Emergence | Postulated Primary Function |
|---|---|---|---|
| Dimer of tRNA-like molecules | Low (Simplest) | Realistically feasible from random RNA chains | Non-coded peptide bond formation and simple elongation |
| Intermediate complexity structures | Medium | Lower statistical likelihood | Catalysis with moderate efficiency |
| Complex symmetrical structures | High | Implausibly low in prebiotic conditions | Advanced catalytic functions |
Prior to the establishment of templated synthesis, short, compositionally biased peptides and RNAs coexisted and interacted through physicochemical interactions driven by environmental availability rather than biosynthetic pathways [130]. Early peptide-RNA interactions likely involved a limited set of "early" amino acids (e.g., Gly, Ala, Asp, Val, Glu, Ile, Leu, Pro, Ser, Thr) [130]. The initial driving forces for these associations would have been:
- Ionic interactions, potentially mediated by prebiotically plausible diamino acids like ornithine (Orn) in the absence of modern basic amino acids (Arg, Lys) [130].
- The gradual replacement of Mg²⁺ ions for charge stabilization of the RNA backbone by short, basic peptides [130].
- The later recruitment of aromatic residues for stacking interactions, which was critical for enhancing binding specificity and stability [130].
The following diagram illustrates the hypothesized transition from prebiotic molecular interactions to the first functional proto-ribosome:
Methodologies for Tracing Ribosomal Accretion
Computational and Phylogenetic Analysis
Reconstructing the ribosome's evolutionary timeline relies on sophisticated computational analyses that treat its structure as a historical document:
- Ideographic (Historical) Framework: This retrodictive approach involves building detailed timelines of structural part accretion. Studies census millions of protein structural domains and molecular functions to construct phylogenetic trees that reveal a 'metabolic-first' origin of proteins, with translation machinery developing later [127] [128].
- Nomothetic (Universal) Framework: This predictive approach seeks universal principles governing macromolecular growth. It examines patterns of coaxial helical stacking and tertiary interactions in rRNA to uncover dynamics of outward and inward ribosomal growth, moving beyond simple concentric layering assumptions [127] [128].
- Network Archaeology: By comparing ribosomal protein networks across bacteria, archaea, and eukaryotes, researchers can identify a universal ancestral core (the ABE network) and trace kingdom-specific developments [131]. This involves meticulous mapping of protein-protein interactions (PPi) and their conservation across evolutionary lineages.
Experimental Validation of Ancient Interactions
Computational predictions require experimental validation through biochemical and biophysical methods:
- Microscale Thermophoresis (MST) and Atomistic Simulations: These techniques are used to investigate the binding of short, ancestral peptide fragments to reconstructed proto-ribosomal RNA (prRNA) constructs. Findings indicate that these peptides bind more specifically to larger, more complex RNA constructs, suggesting that interaction specificity co-evolved with rising architectural complexity [130].
- Conservation Mapping: Systematically comparing interface conservation in ribosomal protein networks across kingdoms reveals that these tiny interfaces are structurally and phylogenetically well conserved, with eukaryotic interfaces showing exceptionally high conservation, particularly in the small subunit [131].
- Structural Analysis of Ancient Fragments: The oldest protein regions within the ribosome are identified by their deep burial within the ribosomal core and their lack of defined secondary structure, suggesting early interactions were flexible and non-specific [130].
Timeline of Ribosomal Accretion and Functional Diversification
The evolutionary timeline of the ribosome reveals a clear trajectory from a simple catalytic core to a complex, allosterically regulated machine.
The Accretion Timeline
Computational studies have reconstructed the following sequence of major events in ribosomal evolution:
Kingdom-Specific Network Expansion
The universal ABE network served as a foundation for distinct evolutionary pathways in different domains of life. The quantitative details of these expansions are summarized in the table below:
Table 2: Quantitative Analysis of Ribosomal Protein Network Expansion Across Kingdoms
| Evolutionary Transition | New Interactions Involving\nNewly Acquired Proteins | New Interactions Involving\nNewly Acquired Extensions | Total New Connections | Key Molecular Characteristics |
|---|---|---|---|---|
| ABE → Bacteria (B) | 77% (58% U-B + 19% B-B) | 21% (12% Ub-U + 9% Ub-B) | Not Specified | Massive incorporation of new bacterial r-proteins |
| ABE → Archaea (A) | 66% (47% U-A + 19% A-A) | 35% (Ua contributions) | Not Specified | Significant role for archaeal-specific extensions |
| Archaea → Eukarya (E) | Minority of new contacts | 57% (Ue + Ae extensions) | Spectacular increase in LSU connectivity | Massive recruitment of aromatic residues for allostery |
The data reveal distinct molecular strategies for network expansion in each domain. Whereas bacteria and archaea extensively incorporated new proteins, eukaryotes predominantly enhanced connectivity through the acquisition of new extensions on existing proteins, particularly optimizing them for allosteric communication [131].
The Rise of Allosteric Networks and Functional Coordination
A pivotal outcome of ribosomal accretion was the development of sophisticated allosteric networks enabling long-range communication between functional centers.
Evolution of the Ribosomal Protein Interactome
The ribosomal protein network evolved into a highly interconnected, non-random graph where r-proteins collectively coevolved to optimize interconnections between functional centers [131]. This network facilitates communication between:
- The decoding center and the Sarcin Ricin Loop (SRL) or E-tRNA site [131].
- The peptidyl transferase center (PTC) and the A-site, mediated by proteins like uL3 [131].
- The peptide exit tunnel and the PTC, allowing regulation of co-translational folding [131].
The architecture of these networks is functionally organized, with r-proteins clustering in modules around main functional centers (mRNA, tRNAs, PTC, peptide tunnel), while others build bridges between these modules or between ribosomal subunits [131].
The Role of Aromatic Residues in Allostery
The eukaryotic evolutionary transition is marked by a massive acquisition of conserved aromatic residues at protein interfaces and along extensions of newly connected r-proteins [131]. This indicates strong selective pressure acting on their sequences, likely for the formation of new allosteric pathways within the network. These aromatic residues facilitate allosteric information transfer that coordinates ribosomal dynamics during tRNA translocation and association with translation factors [130].
The Scientist's Toolkit: Key Research Reagents and Methods
Table 3: Essential Research Reagents and Methodologies for Studying Ribosomal Evolution
| Reagent / Method | Category | Function / Application | Key Insight Enabled |
|---|---|---|---|
| Proto-ribosomal RNA (prRNA) Constructs | RNA Preparation | Small (136nt) and large (617nt) constructs for binding studies with ancestral peptide fragments [130] | Demonstrated increased binding specificity with rising RNA complexity |
| Microscale Thermophoresis (MST) | Biophysical Assay | Quantifies binding affinity and kinetics in peptide-RNA interactions [130] | Validated co-evolution of specificity with architectural complexity |
| Atomistic Computer Simulations | Computational Modeling | Models molecular interactions at atomic-level resolution [130] | Revealed dynamics of early peptide-RNA binding |
| Phylogenetic Trees of Protein Domains | Computational Analysis | Census of millions of protein domains to reconstruct evolutionary timelines [127] [128] | Revealed 'metabolic-first' protein origin, late translation development |
| Ribosomal Protein Network Maps | Structural Bioinformatics | Comparative analysis of protein-protein interactions across kingdoms [131] | Identified universal ABE core and kingdom-specific adaptations |
The principle of evolutionary accretion provides a powerful framework for understanding the ribosome's layered architecture and its emergence from the prebiotic world. The evidence confirms that the ribosome grew from a simple, symmetrical proto-ribosome through the sequential addition of RNA and protein components, driven by the selective advantage of enhanced functionality and allosteric control. This process created a sophisticated molecular machine capable of precise information transfer and coordination.
Future research directions should focus on:
- Synthetic Biology Applications: Reconstructing intermediate ancestral ribosome states to test evolutionary hypotheses and engineer novel functionalities [127].
- Expanding the Toolkit: Developing more sophisticated experimental and computational methods to probe earlier stages of ribosomal evolution, particularly the transition from unstructured peptides to structured protein components.
- Allosteric Pathway Engineering: Leveraging insights from ribosomal allostery to design new biomolecular machines with engineered communication networks.
The ribosome stands as a testament to the incremental power of evolutionary processes, transforming simple molecular interactions into one of life's most essential and complex macromolecular machines.
Conclusion
The RNA World hypothesis has evolved from a compelling idea into a robust, testable framework supported by recent chemical breakthroughs, ancient biosignature detection, and laboratory evolution of functional RNAs. The discovery that RNA can spontaneously connect with amino acids via thioester intermediates under prebiotic conditions provides a plausible path for the transition from an RNA world to the RNA-protein world that characterizes all modern life. For biomedical researchers and drug developers, these foundational insights are now driving a therapeutic revolution. RNA's ancient capacity for information storage and catalysis is being harnessed in mRNA vaccines, RNA-targeting small molecules, and synthetic biological systems. Future directions will focus on achieving autonomous self-replicating RNA systems, expanding the functional repertoire of artificial ribozymes, and leveraging AI to accelerate RNA therapeutic design. Understanding life's origins is not just about explaining the past—it's providing a blueprint for the future of medicine.
References
- 1. - The School of Biomedical Sciences Wiki RNA world hypothesis [https://teaching.ncl.ac.uk/bms/wiki/index.php/RNA_world_hypothesis]
- 2. : What We Know, What We Don't | Evolution... Prebiotic Chemistry [https://evolution-outreach.biomedcentral.com/articles/10.1007/s12052-012-0443-9]
- 3. : Origin of Life Explained Simply RNA World Hypothesis [https://www.vedantu.com/biology/rna-world-hypothesis]
- 4. What is the RNA ? World Hypothesis [https://www.news-medical.net/life-sciences/What-is-the-RNA-World-Hypothesis.aspx]
- 5. RNA Structure - Function Protocols: Alkaline Hydrolysis, RNA Sequencing and RNA Structure Analyses with Nucleases | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/references/protocols/nucleic-acid-purification-and-analysis/rna-protocol/rna-structure-function-protocols.html]
- 6. Advances and opportunities in RNA structure experimental determination and computational modeling | Nature Methods [https://www.nature.com/articles/s41592-022-01623-y]
- 7. High-Throughput Determination of RNA Structures - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7388734/]
- 8. The RNA : the worst theory of the early evolution of... world hypothesis [https://pubmed.ncbi.nlm.nih.gov/22793875/]
- 9. Rethinking " Prebiotic " Chemistry [https://authors.library.caltech.edu/records/2kmp7-p5355]
- 10. The RNA world 'hypothesis' [https://www.nature.com/articles/s41580-022-00514-6]
- 11. Prebiotic chemistry and the origin of the RNA world - PubMed [https://pubmed.ncbi.nlm.nih.gov/15217990/]
- 12. RNA's hidden potential: New study unveils its role in early life and future bioengineering | ScienceDaily [https://www.sciencedaily.com/releases/2024/04/240418111806.htm]
- 13. A molecular of the RNA world - Ars Technica fossil [https://arstechnica.com/science/2011/07/a-molecular-fossil-of-the-rna-world/]
- 14. Could a Proto-Ribosome Emerge Spontaneously in the Prebiotic World? - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6274258/]
- 15. Peeling the onion: ribosomes are ancient molecular fossils [https://pubmed.ncbi.nlm.nih.gov/19628620/]
- 16. as Ancient Ribosomes | News | Astrobiology Molecular Fossils [https://astrobiology.nasa.gov/news/ribosomes-as-ancient-molecular-fossils/]
- 17. (PDF) Peeling the Onion: Ribosomes Are Ancient Molecular Fossils [https://www.academia.edu/128800798/Peeling_the_Onion_Ribosomes_Are_Ancient_Molecular_Fossils]
- 18. as First RNA Material (1.3.6) | IB DP Biology HL... | TutorChase Genetic [https://www.tutorchase.com/notes/ib/biology-2025-hl/1-3-6-rna-as-first-genetic-material]
- 19. 's Role in Life's Origins - RNA 's Vital RNA Dual Functions [https://scisimple.com/en/articles/2025-10-04-rnas-role-in-lifes-origins--ak2zz81]
- 20. Dynamics of prebiotic RNA reproduction illuminated by chemical game theory - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4983821/]
- 21. Prebiotic Chemistry [https://www.cup.uni-muenchen.de/oc/trapp/prebiotic.html]
- 22. The Incredible Conundrum of Life's Origin - Nautilus [https://nautil.us/the-incredible-conundrum-of-lifes-origin-1178890/]
- 23. The RNA : Origins and Implications World [https://www.numberanalytics.com/blog/rna-world-origins-astrobiology]
- 24. The RNA world hypothesis: the worst theory of the early evolution of... [https://biologydirect.biomedcentral.com/articles/10.1186/1745-6150-7-23]
- 25. The sugar that sparked life: Why ribose was RNA ’s first... | ScienceDaily [https://www.sciencedaily.com/releases/2025/07/250722035558.htm]
- 26. Scientists Solved a Key Mystery Regarding the Evolution of Life on Earth [https://www.zmescience.com/research/mystery-origin-of-life-proteins-rna-dna/]
- 27. The Origin of Prebiotic Information System in the Peptide/RNA World: A Simulation Model of the Evolution of Translation and the Genetic Code - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6463137/]
- 28. Scientists recreate life’s first step: Linking amino acids to RNA | ScienceDaily [https://www.sciencedaily.com/releases/2025/08/250828002406.htm]
- 29. The Molecular Architect: RNA, Amino Acids, and the Self-Assembling Ribozyme | Department of Chemistry | The University of Chicago [https://chemistry.uchicago.edu/news/molecular-architect-rna-amino-acids-and-self-assembling-ribozyme]
- 30. Amino acids catalyse RNA formation under ambient alkaline conditions | Nature Communications [https://www.nature.com/articles/s41467-025-60359-3]
- 31. Spontaneous Formation of RNA Strands, Peptidyl RNA, and Cofactors - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4678511/]
- 32. 学 RNA - 维基百科,自由的百科全书 世 界 说 [https://zh.wikipedia.org/zh-sg/RNA%E4%B8%96%E7%95%8C%E5%AD%B8%E8%AA%AA]
- 33. Experiment sheds light on the origin of life, supporting the existence of a ‘thioester world’ before living beings | Science | EL PAÍS English [https://english.elpais.com/science-tech/2025-08-27/experiment-sheds-light-on-the-origin-of-life-supporting-the-existence-of-a-thioester-world-before-living-beings.html]
- 34. Scientists Think They’ve Found Life’s Original Catalyst [https://www.popularmechanics.com/science/a65940591/chemical-reaction-create-life/]
- 35. Thioester-mediated RNA aminoacylation and peptidyl-RNA synthesis in water | Nature [https://www.nature.com/articles/s41586-025-09388-y]
- 36. Chemists Have Replicated a Critical Moment in The... : ScienceAlert [https://www.sciencealert.com/chemists-have-recreated-a-critical-moment-in-the-creation-of-life]
- 37. RNA and Sulfur Compounds Possibly Created the First Peptides on Earth | The Scientist [https://www.the-scientist.com/rna-and-sulfur-compounds-possibly-created-the-first-peptides-on-earth-73323]
- 38. RNA world - Wikipedia [https://en.wikipedia.org/wiki/RNA_world]
- 39. Witnessing the structural evolution of an RNA enzyme | eLife [https://elifesciences.org/articles/71557]
- 40. -catalysed Ribozyme synthesis using triplet building blocks | eLife RNA [https://elifesciences.org/articles/35255]
- 41. - Wikipedia Ribozyme [https://en.wikipedia.org/wiki/Ribozyme]
- 42. Rolling circle RNA synthesis catalyzed by RNA | eLife [https://elifesciences.org/articles/75186]
- 43. A structural analysis of in vitro catalytic activities of hammerhead... [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-8-469]
- 44. : Unraveling the mysteries of life's origins - Earth.com RNA World [https://www.earth.com/news/rna-world-unraveling-mysteries-life-origin/]
- 45. Zhi John Lu’s team published an AI , RNAsmol, to model ... predict [https://life.tsinghua.edu.cn/lifeen/info/1131/1902.htm]
- 46. Artificial Intelligence in Small-Molecule Drug Discovery: A Critical Review of Methods, Applications, and Real-World Outcomes - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12472608/]
- 47. Deep dive into RNA: a systematic literature review on RNA structure prediction using machine learning methods | Artificial Intelligence Review [https://link.springer.com/article/10.1007/s10462-024-10910-3]
- 48. From Sequence to Structure: A Fast Track for RNA Modeling - Berkeley Lab – Berkeley Lab News Center [https://newscenter.lbl.gov/2025/05/13/from-sequence-to-structure-a-fast-track-for-rna-modeling/]
- 49. Dynamics and RNA Revealed through Atomistic... Interactions [https://arxiv.org/html/2507.08474v1]
- 50. The prediction of RNA - small binding sites in molecule ... RNA [https://pubmed.ncbi.nlm.nih.gov/40268011/]
- 51. AI Models Enhance the Drug Discovery Portfolio [https://www.genengnews.com/topics/artificial-intelligence/ai-models-enhance-the-drug-discovery-portfolio/]
- 52. The non-canonical nucleotides and prebiotic evolution [https://pubmed.ncbi.nlm.nih.gov/39900260/]
- 53. Within an Synthetic Central Dogma... Biological Circuits Orthogonal [https://pmc.ncbi.nlm.nih.gov/articles/PMC7746572/]
- 54. Design and Evaluation of Synthetic -Based Incoherent... RNA [https://pmc.ncbi.nlm.nih.gov/articles/PMC8391864/]
- 55. for Biological : Berkeley Lab Researchers... Circuits Synthetic Biology [https://newscenter.lbl.gov/2011/05/26/biological-circuits-for-synthetic-biology/]
- 56. Publications | Research Activities | CiRA | Center for iPS Cell Research... [https://www.cira.kyoto-u.ac.jp/e/research/finding/200311-150000.html]
- 57. Synthetic spike-in standards for RNA-seq experiments - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3166838/]
- 58. Targeting RNA splicing modulation: new perspectives for anticancer strategy? | Journal of Experimental & Clinical Cancer Research | Full Text [https://jeccr.biomedcentral.com/articles/10.1186/s13046-025-03279-w]
- 59. The growth of siRNA-based therapeutics: updated clinical studies - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8187268/]
- 60. Overcoming delivery barriers to unlock the full potential of RNA -based... [https://www.iptonline.com/articles/overcoming-delivery-barriers-to-unlock-the-full-potential-of-rna-based-medicine-spring-2025]
- 61. Frontiers | Clinical development prospects of siRNA drugs for tumor therapy: analysis of clinical trial registration data from 2004 to 2024 [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1637958/full]
- 62. Splicing modulators: on the way from nature to clinic | The Journal of Antibiotics [https://www.nature.com/articles/s41429-021-00450-1]
- 63. Recent advances in RNA - targeting therapy for neurological diseases... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10358688/]
- 64. RNA Therapeutics Market is expected to generate a revenue of USD 22.37 Billion by 2032, Globally, at 9.4% CAGR: Verified Market Research® [https://www.globenewswire.com/news-release/2025/03/17/3043838/0/en/RNA-Therapeutics-Market-is-expected-to-generate-a-revenue-of-USD-22-37-Billion-by-2032-Globally-at-9-4-CAGR-Verified-Market-Research.html]
- 65. RNA Therapeutics Market Size, Share & 2030 Trends Report [https://www.mordorintelligence.com/industry-reports/rna-therapeutics-market]
- 66. The difficult case of an RNA -only origin of life - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7289000/]
- 67. in the Real World: 5 Uses You'll Actually See ( Thioesters ) 2025 [https://www.linkedin.com/pulse/thioesters-real-world-5-uses-youll-actually-see-2025-yxnfc]
- 68. Support Efficient Protein Biosynthesis by the Ribosome Thioesters [https://pubmed.ncbi.nlm.nih.gov/40161951/]
- 69. Insights into the Mechanism and Catalysis of Peptide ... Thioester [https://pmc.ncbi.nlm.nih.gov/articles/PMC7961367/]
- 70. An enzyme for high-yield, ATP-driven C-terminal thioester generation | Nature Chemistry [https://www.nature.com/articles/s41557-025-01872-2]
- 71. On the origin of life: an RNA -focused synthesis and narrative - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10351881/]
- 72. Frontiers | mRNA : linking infectious disease... vaccine platforms [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2025.1547025/full]
- 73. Clinical development of therapeutic mRNA applications - Icahn School of Medicine at Mount Sinai [https://scholars.mssm.edu/en/publications/clinical-development-of-therapeutic-mrna-applications]
- 74. mRNA in the Context of Protein Replacement Therapy [https://www.mdpi.com/1999-4923/15/1/166]
- 75. Analytical Strategies for Current -Based mRNA - PMC Therapeutics [https://pmc.ncbi.nlm.nih.gov/articles/PMC11990077/]
- 76. Lipid nanoparticles for mRNA therapy: recent advances in targeted delivery - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11749733/]
- 77. mRNA Therapeutics Market Size & Share 2025-2030 [https://www.360iresearch.com/library/intelligence/mrna-therapeutics]
- 78. Market: Trends, Size, and Global Outlook mRNA Therapeutics [https://blog.bccresearch.com/mrna-therapeutics-market-trends-size-and-global-outlook]
- 79. Non-enzymatic primer extension with strand displacement | eLife [https://elifesciences.org/articles/51888]
- 80. pH-Driven RNA under Prebiotically Plausible... Strand Separation [https://pubmed.ncbi.nlm.nih.gov/30383375/]
- 81. The RNA World and the Origins of Life - Molecular Biology of the Cell - NCBI Bookshelf [https://www.ncbi.nlm.nih.gov/books/NBK26876/]
- 82. The RNA world hypothesis: the worst theory of the early evolution of life (except for all the others)a - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3495036/]
- 83. The RNA i-Motif in the Primordial RNA World [https://pubmed.ncbi.nlm.nih.gov/31127498/]
- 84. The Origins of the RNA World - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3331698/]
- 85. Metals in Prebiotic : A Possible Evolutionary Pathway for the... Catalysis [https://pmc.ncbi.nlm.nih.gov/articles/PMC9933472/]
- 86. Virus RNA Mutants: A Useful Tool for Evolutionary Biology or... Fidelity [https://pmc.ncbi.nlm.nih.gov/articles/PMC6267201/]
- 87. A pro-metastatic tRNA fragment drives Nucleolin oligomerization and... [https://pubmed.ncbi.nlm.nih.gov/35654044/]
- 88. Nucleotide-induced hyper- oligomerization ... | Nature Communications [https://www.nature.com/articles/s41467-025-56824-8?error=cookies_not_supported&code=780cdba6-1a60-4b83-9eab-d48487c3b277]
- 89. Discovery of RNA‐Targeting Small Molecules: Challenges and Future Directions - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12375692/]
- 90. Unlocking the regulatory code of RNA: launching the Human RNome Project | Genome Biology | Full Text [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-025-03824-y]
- 91. pH-Dependent peptide bond formation by the selective of... coupling [https://pmc.ncbi.nlm.nih.gov/articles/PMC7808311/]
- 92. Amino acid analogues provide multiple plausible pathways to prebiotic peptides - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11077012/]
- 93. A prebiotically plausible scenario of an RNA–peptide world | Nature [https://www.nature.com/articles/s41586-022-04676-3]
- 94. Prebiotic Chemistry: Origin & Techniques | Vaia [https://www.vaia.com/en-us/explanations/physics/astrophysics/prebiotic-chemistry/]
- 95. 生命起源假说:RNA-多肽世界的前生物图景 | 集智俱乐部 [https://swarma.org/?p=35736]
- 96. AI decodes ancient 'whispers' of life in 3.3 billion-year-old rocks [https://interestingengineering.com/science/3-billion-year-old-rocks]
- 97. Noncanonical RNA Nucleosides as Molecular Fossils of an Early... [https://pubmed.ncbi.nlm.nih.gov/29533524/]
- 98. report earliest molecular evidence of photosynthetic life Researchers [https://cen.acs.org/physical-chemistry/astrochemistry/Researchers-report-earliest-molecular-evidence/103/web/2025/11]
- 99. New method for analysing fossils | Interviews | Naked Scientists [https://www.thenakedscientists.com/articles/interviews/using-machine-learning-analyse-fossils]
- 100. Knowledge-aware contrastive heterogeneous molecular graph learning | PLOS Computational Biology [https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1013008]
- 101. MolParser: End-to-end Visual Recognition of Molecule Structures in the Wild [https://arxiv.org/html/2411.11098v1]
- 102. and DNA : What Makes Them Different? | Technology Networks RNA [https://www.technologynetworks.com/genomics/articles/what-are-the-key-differences-between-dna-and-rna-296719]
- 103. Structure and function [https://www.asbmb.org/education/core-concept-teaching-strategies/foundational-concepts/structure-function]
- 104. The protometabolic nature of prebiotic chemistry - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10614573/]
- 105. Prebiotic chemistry: a new modus operandi - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3158916/]
- 106. Prebiotic Chemistry Experiments Using Microfluidic Devices - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9605377/]
- 107. Correlations between RNA and protein expression profiles in 23 human cell lines | BMC Genomics | Full Text [https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-10-365]
- 108. Simultaneous quantification of mRNA and protein in single cells reveals post-transcriptional effects of genetic variation | eLife [https://elifesciences.org/articles/60645]
- 109. and optimization of methods for the simultaneous... Comparison [https://pubmed.ncbi.nlm.nih.gov/27237373/]
- 110. RNA - Wikipedia [https://en.wikipedia.org/wiki/RNA]
- 111. of Synthesis by In Vitro Transcription | SpringerLink RNA [https://link.springer.com/protocol/10.1007/978-1-59745-248-9_3]
- 112. Rapid enzymatic synthesis of long RNA polymers: A simple protocol ... [https://pubmed.ncbi.nlm.nih.gov/30926532/]
- 113. Construction of RNA reference materials for improving the quantification of transcriptomic data - PubMed [https://pubmed.ncbi.nlm.nih.gov/39966680/]
- 114. Paleobiological Perspectives on Early Microbial Evolution - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4484972/]
- 115. Stromatolite - Wikipedia [https://en.wikipedia.org/wiki/Stromatolite]
- 116. Stromatolite - an overview | ScienceDirect Topics [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/stromatolite]
- 117. Chemical evidence of ancient life detected in 3.3- billion - year - old rocks [https://carnegiescience.edu/chemical-evidence-ancient-life-detected-33-billion-year-old-rocks]
- 118. Signs Of Photosynthesis Identified In 2 . 5 - Billion - Year ... Old Rocks [https://www.iflscience.com/traces-of-photosynthetic-lifeforms-1-billion-years-older-than-previous-record-holder-discovered-81588]
- 119. Ancient Life Discovery: Chemical Traces Push Origins Back 800 Million... [https://www.downtoearth.org.in/science-technology/chemical-whispers-reveal-life-began-800-million-years-earlier-than-thought]
- 120. Before DNA, before RNA: Life in the hodge-podge world | New Scientist [https://www.newscientist.com/article/dn21335-before-dna-before-rna-life-in-the-hodge-podge-world/]
- 121. Peptide nucleic acid | PPTX [https://www.slideshare.net/slideshow/peptide-nucleic-acid-43835359/43835359]
- 122. Acyclic (S)-glycol nucleic acid (S-GNA) modification of siRNAs improves the safety of RNAi therapeutics while maintaining potency - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10019370/]
- 123. Experimental evidence that GNA and TNA were not sequential polymers in the prebiotic evolution of RNA - PubMed [https://pubmed.ncbi.nlm.nih.gov/17828568/]
- 124. Life as a guide to prebiotic nucleotide synthesis | Nature Communications [https://www.nature.com/articles/s41467-018-07220-y]
- 125. Prebiotic Synthesis of RNA Nucleotides - Astrobiology [https://astrobiology.com/2018/01/prebiotic-synthesis-of-rna-nucleotides.html]
- 126. and Early Evolution of the Origins | SpringerLink Ribosome [https://link.springer.com/chapter/10.1007/978-3-319-39468-8_3]
- 127. Computing the origin and evolution of the ribosome from its structure - Uncovering processes of macromolecular accretion benefiting synthetic biology - PubMed [https://pubmed.ncbi.nlm.nih.gov/27096056/]
- 128. Computing the origin and evolution of the ribosome from its structure — Uncovering processes of macromolecular accretion benefiting synthetic biology - ScienceDirect [https://www.sciencedirect.com/science/article/pii/S2001037015000355]
- 129. Could a Proto- Ribosome Emerge Spontaneously in the Prebiotic ... [https://pubmed.ncbi.nlm.nih.gov/27941673/]
- 130. Evolution of protein–RNA interactions [https://arxiv.org/html/2505.02037]
- 131. of Evolution protein network ribosomal | Scientific... architectures [https://www.nature.com/articles/s41598-020-80194-4?error=cookies_not_supported&code=8cd9dcd8-5ed6-4800-8a01-fbf4d0f6d312]