Research Articles
Solving Molecular Dynamics Equilibration: A Practical Guide to Temperature and Pressure Control for Reliable Simulations
Achieving proper temperature and pressure equilibration in Molecular Dynamics (MD) simulations is a critical yet often challenging step for obtaining physically meaningful results in biomedical research, particularly in drug development.
Molecular Dynamics Simulation Crashes: A Comprehensive Guide from Prevention to Recovery for Biomedical Researchers
This article provides a systematic framework for researchers and drug development professionals to handle molecular dynamics simulation crashes.
Accelerate Your Research: A Practical Guide to Optimizing Slow Molecular Dynamics Simulations
Molecular dynamics (MD) simulations are a cornerstone of computational chemistry, biophysics, and drug discovery, yet their extreme computational cost often hinders research progress.
Solving Molecular Dynamics Energy Minimization Failures: A Troubleshooting Guide for Computational Researchers
This article provides a comprehensive framework for understanding, troubleshooting, and resolving common failures in molecular dynamics (MD) energy minimization.
Avoiding Common Pitfalls in Molecular Dynamics Simulations: A Guide to Error Correction, Force Field Selection, and Best Practices
This article provides a comprehensive guide for researchers and drug development professionals on identifying, troubleshooting, and preventing common errors in molecular dynamics (MD) simulations.
How to Fix Molecular Dynamics System Instability: A Troubleshooting Guide for Researchers
Molecular dynamics (MD) simulations are powerful but prone to instability that can invalidate results and waste computational resources.
Root Mean Square Deviation (RMSD) in Molecular Dynamics: A Comprehensive Guide for Biomolecular Research and Drug Discovery
Root Mean Square Deviation (RMSD) is a fundamental metric in molecular dynamics (MD) simulations, providing critical insights into biomolecular structural stability, conformational changes, and ligand-binding interactions.
A Practical Guide to Explicit Solvent Molecular Dynamics for Drug Discovery and Biomolecular Research
This comprehensive guide provides researchers and drug development professionals with foundational knowledge and advanced methodologies for performing molecular dynamics simulations with explicit solvent.
From Folding Pathways to Drug Discovery: Leveraging Molecular Dynamics for Protein Simulation
This article provides a comprehensive overview of how Molecular Dynamics (MD) simulations have revolutionized the study of protein folding, moving beyond static structures to capture dynamic conformational ensembles.
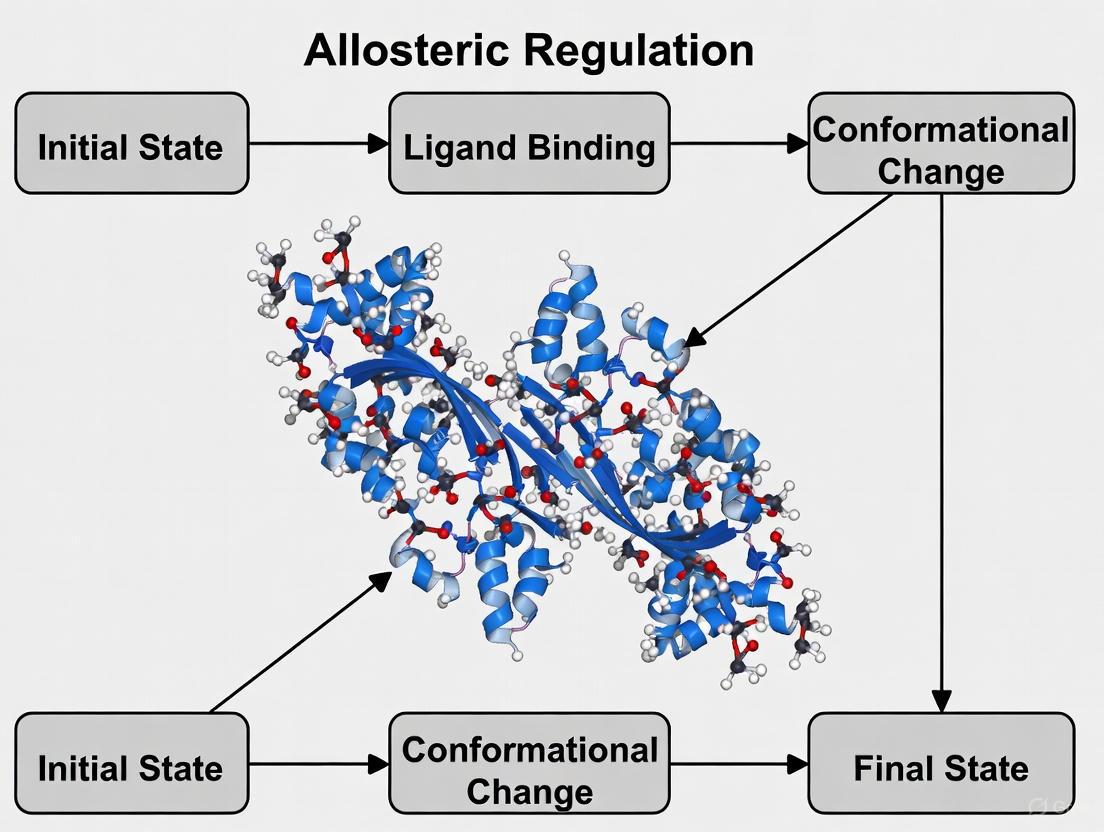
Decoding Allostery: How Molecular Dynamics Simulations Are Revolutionizing Drug Discovery
Allosteric regulation, the process of controlling protein function through binding at distal sites, offers a promising avenue for developing highly selective therapeutics.